bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0393_orf1
Length=183
Score E
Sequences producing significant alignments: (Bits) Value
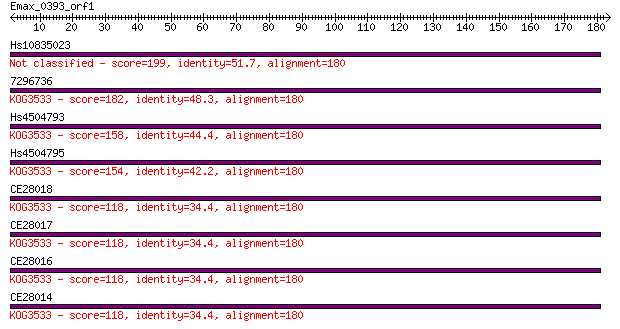
Hs10835023 199 3e-51
7296736 182 3e-46
Hs4504793 158 7e-39
Hs4504795 154 7e-38
CE28018 118 7e-27
CE28017 118 7e-27
CE28016 118 7e-27
CE28014 118 7e-27
> Hs10835023
Length=2695
Score = 199 bits (506), Expect = 3e-51, Method: Composition-based stats.
Identities = 93/181 (51%), Positives = 124/181 (68%), Gaps = 1/181 (0%)
Query 1 QGSKVLDFFERTEDLFGEMNWQKRLREQPLLYWVSRHMSLWNSFSFYLAFVVNLIVAFFY 60
QGSK+ DFF R+EDLF EMNWQK+LR QP+LYW +R+MS W+S SF LA ++NL+VAFFY
Sbjct 2181 QGSKINDFFLRSEDLFNEMNWQKKLRAQPVLYWCARNMSFWSSISFNLAVLMNLLVAFFY 2240
Query 61 PFPA-KAVEIDGRISAAIWAILFMSLAAVFAFPGKLGFRVLIGSALVRLIYSIDIQPTLW 119
PF + ++ S +W + +SLA V A P G R LI S ++RLI+S+ +QPTL+
Sbjct 2241 PFKGVRGGTLEPHWSGLLWTAMLISLAIVIALPKPHGIRALIASTILRLIFSVGLQPTLF 2300
Query 120 LLGTTQVVIKAIHLISIMGNHGTFNKSISRILIDREFLYHINYLFFCVIGLCCHPFCYSV 179
LLG V K I L+S +GN GTF + +++D EFLYH+ YL C +GL H F YS+
Sbjct 2301 LLGAFNVCNKIIFLMSFVGNCGTFTRGYRAMVLDVEFLYHLLYLVICAMGLFVHEFFYSL 2360
Query 180 L 180
L
Sbjct 2361 L 2361
> 7296736
Length=2837
Score = 182 bits (462), Expect = 3e-46, Method: Compositional matrix adjust.
Identities = 87/180 (48%), Positives = 117/180 (65%), Gaps = 0/180 (0%)
Query 1 QGSKVLDFFERTEDLFGEMNWQKRLREQPLLYWVSRHMSLWNSFSFYLAFVVNLIVAFFY 60
QGSKV DFF++ E++F EM WQK+LR QPLL+W+S +MSLW++ F V+N+IVAFFY
Sbjct 2328 QGSKVADFFDKAEEMFNEMKWQKKLRSQPLLFWISSYMSLWSNILFNCVVVINMIVAFFY 2387
Query 61 PFPAKAVEIDGRISAAIWAILFMSLAAVFAFPGKLGFRVLIGSALVRLIYSIDIQPTLWL 120
PF E+ IS W I SL V A P + G R IGS ++R I+ + + TL L
Sbjct 2388 PFDNTVPELSSHISLLFWIITIFSLVIVLALPRESGIRTFIGSVILRFIFLLGPESTLCL 2447
Query 121 LGTTQVVIKAIHLISIMGNHGTFNKSISRILIDREFLYHINYLFFCVIGLCCHPFCYSVL 180
LG V +K++H++SIMGN GT K + +I+ D + LYH Y+ FC GL HPF YS+L
Sbjct 2448 LGVVTVTLKSVHIVSIMGNKGTLEKQLIKIITDFQLLYHCIYIAFCFCGLIFHPFFYSLL 2507
> Hs4504793
Length=2701
Score = 158 bits (399), Expect = 7e-39, Method: Compositional matrix adjust.
Identities = 80/182 (43%), Positives = 112/182 (61%), Gaps = 2/182 (1%)
Query 1 QGSKVLDFFERTEDLFGEMNWQKRLREQPLLYWVSRHMSLWNSFSFYLAFVVNLIVAFFY 60
QGSKV DFF++TEDL+ EM WQK++R P L+W SRH+SLW S SF LA +NL VA FY
Sbjct 2189 QGSKVNDFFQQTEDLYNEMKWQKKIRNNPALFWFSRHISLWGSISFNLAVFINLAVALFY 2248
Query 61 PFPAKAVE--IDGRISAAIWAILFMSLAAVFAFPGKLGFRVLIGSALVRLIYSIDIQPTL 118
PF E + S +W + + + +F F +G R + S ++R IY+I + PTL
Sbjct 2249 PFGDDGDEGTLSPLFSVLLWIAVAICTSMLFFFSKPVGIRPFLVSIMLRSIYTIGLGPTL 2308
Query 119 WLLGTTQVVIKAIHLISIMGNHGTFNKSISRILIDREFLYHINYLFFCVIGLCCHPFCYS 178
LLG + K + L+S +GN GTF + +++D FLYH+ Y+ C++GL H F YS
Sbjct 2309 ILLGAANLCNKIVFLVSFVGNRGTFTRGYRAVILDMAFLYHVAYVLVCMLGLFVHEFFYS 2368
Query 179 VL 180
L
Sbjct 2369 FL 2370
> Hs4504795
Length=2671
Score = 154 bits (390), Expect = 7e-38, Method: Composition-based stats.
Identities = 76/183 (41%), Positives = 111/183 (60%), Gaps = 5/183 (2%)
Query 1 QGSKVLDFFERTEDLFGEMNWQKRLREQPLLYWVSRHMSLWNSFSFYLAFVVNLIVAFFY 60
QGSKV DFF+++ L EM WQ+ +R PL+YW SR M+LW S SF LA +N+I+AFFY
Sbjct 2164 QGSKVSDFFDQSSFLHNEMEWQRNVRSMPLIYWFSRRMTLWGSISFNLAVFINIIIAFFY 2223
Query 61 PF---PAKAVEIDGRISAAIWAILFMSLAAVFAFPGKLGFRVLIGSALVRLIYSIDIQPT 117
P+ + V IS W ++ S+AA+F + R LI + ++R IY + I PT
Sbjct 2224 PYMEGASTGVLDSPLISLLFWILICFSIAALFT--KRYSIRPLIVALILRSIYYLGIGPT 2281
Query 118 LWLLGTTQVVIKAIHLISIMGNHGTFNKSISRILIDREFLYHINYLFFCVIGLCCHPFCY 177
L +LG + K + ++S +GN GTF + +++D EFLYH+ Y+ V+GL H Y
Sbjct 2282 LNILGALNLTNKIVFVVSFVGNRGTFIRGYKAMVMDMEFLYHVGYILTSVLGLFAHELFY 2341
Query 178 SVL 180
S+L
Sbjct 2342 SIL 2344
> CE28018
Length=2846
Score = 118 bits (295), Expect = 7e-27, Method: Composition-based stats.
Identities = 62/185 (33%), Positives = 102/185 (55%), Gaps = 5/185 (2%)
Query 1 QGSKVLDFFERTEDLFGEMNWQKRLREQPLLYWVSRHMSLWNSFSFYLAFVVNLIVAFFY 60
QGSKV +FF+ E ++ EM WQ++L+++ L W + + LW SF+ AF+VN +VA +Y
Sbjct 2361 QGSKVTEFFDEWETMYHEMIWQRKLQDRKWLSWCAFRLPLWTRLSFHFAFIVNALVARYY 2420
Query 61 PFPAKAVEIDGRISAAIWAILFMS-LAAVFAFPGKLGFR----VLIGSALVRLIYSIDIQ 115
P P + + W +F S L A + K+ +++ S L+ SI +
Sbjct 2421 PLPEHSNSSISLGNLYSWFAVFSSFLLAHYLRHDKIYLHKTSLLILASLCFLLLSSIGVT 2480
Query 116 PTLWLLGTTQVVIKAIHLISIMGNHGTFNKSISRILIDREFLYHINYLFFCVIGLCCHPF 175
TL++ G Q+V K +H+++ + N G ++ I+ IL R Y + YLF C++GL HP
Sbjct 2481 LTLYIFGILQLVNKIVHVVAFVSNKGLEDRPIAEILACRNLHYLLVYLFICILGLLVHPM 2540
Query 176 CYSVL 180
Y +L
Sbjct 2541 IYCIL 2545
> CE28017
Length=2857
Score = 118 bits (295), Expect = 7e-27, Method: Composition-based stats.
Identities = 62/185 (33%), Positives = 102/185 (55%), Gaps = 5/185 (2%)
Query 1 QGSKVLDFFERTEDLFGEMNWQKRLREQPLLYWVSRHMSLWNSFSFYLAFVVNLIVAFFY 60
QGSKV +FF+ E ++ EM WQ++L+++ L W + + LW SF+ AF+VN +VA +Y
Sbjct 2372 QGSKVTEFFDEWETMYHEMIWQRKLQDRKWLSWCAFRLPLWTRLSFHFAFIVNALVARYY 2431
Query 61 PFPAKAVEIDGRISAAIWAILFMS-LAAVFAFPGKLGFR----VLIGSALVRLIYSIDIQ 115
P P + + W +F S L A + K+ +++ S L+ SI +
Sbjct 2432 PLPEHSNSSISLGNLYSWFAVFSSFLLAHYLRHDKIYLHKTSLLILASLCFLLLSSIGVT 2491
Query 116 PTLWLLGTTQVVIKAIHLISIMGNHGTFNKSISRILIDREFLYHINYLFFCVIGLCCHPF 175
TL++ G Q+V K +H+++ + N G ++ I+ IL R Y + YLF C++GL HP
Sbjct 2492 LTLYIFGILQLVNKIVHVVAFVSNKGLEDRPIAEILACRNLHYLLVYLFICILGLLVHPM 2551
Query 176 CYSVL 180
Y +L
Sbjct 2552 IYCIL 2556
> CE28016
Length=2847
Score = 118 bits (295), Expect = 7e-27, Method: Composition-based stats.
Identities = 62/185 (33%), Positives = 102/185 (55%), Gaps = 5/185 (2%)
Query 1 QGSKVLDFFERTEDLFGEMNWQKRLREQPLLYWVSRHMSLWNSFSFYLAFVVNLIVAFFY 60
QGSKV +FF+ E ++ EM WQ++L+++ L W + + LW SF+ AF+VN +VA +Y
Sbjct 2362 QGSKVTEFFDEWETMYHEMIWQRKLQDRKWLSWCAFRLPLWTRLSFHFAFIVNALVARYY 2421
Query 61 PFPAKAVEIDGRISAAIWAILFMS-LAAVFAFPGKLGFR----VLIGSALVRLIYSIDIQ 115
P P + + W +F S L A + K+ +++ S L+ SI +
Sbjct 2422 PLPEHSNSSISLGNLYSWFAVFSSFLLAHYLRHDKIYLHKTSLLILASLCFLLLSSIGVT 2481
Query 116 PTLWLLGTTQVVIKAIHLISIMGNHGTFNKSISRILIDREFLYHINYLFFCVIGLCCHPF 175
TL++ G Q+V K +H+++ + N G ++ I+ IL R Y + YLF C++GL HP
Sbjct 2482 LTLYIFGILQLVNKIVHVVAFVSNKGLEDRPIAEILACRNLHYLLVYLFICILGLLVHPM 2541
Query 176 CYSVL 180
Y +L
Sbjct 2542 IYCIL 2546
> CE28014
Length=2903
Score = 118 bits (295), Expect = 7e-27, Method: Composition-based stats.
Identities = 62/185 (33%), Positives = 102/185 (55%), Gaps = 5/185 (2%)
Query 1 QGSKVLDFFERTEDLFGEMNWQKRLREQPLLYWVSRHMSLWNSFSFYLAFVVNLIVAFFY 60
QGSKV +FF+ E ++ EM WQ++L+++ L W + + LW SF+ AF+VN +VA +Y
Sbjct 2418 QGSKVTEFFDEWETMYHEMIWQRKLQDRKWLSWCAFRLPLWTRLSFHFAFIVNALVARYY 2477
Query 61 PFPAKAVEIDGRISAAIWAILFMS-LAAVFAFPGKLGFR----VLIGSALVRLIYSIDIQ 115
P P + + W +F S L A + K+ +++ S L+ SI +
Sbjct 2478 PLPEHSNSSISLGNLYSWFAVFSSFLLAHYLRHDKIYLHKTSLLILASLCFLLLSSIGVT 2537
Query 116 PTLWLLGTTQVVIKAIHLISIMGNHGTFNKSISRILIDREFLYHINYLFFCVIGLCCHPF 175
TL++ G Q+V K +H+++ + N G ++ I+ IL R Y + YLF C++GL HP
Sbjct 2538 LTLYIFGILQLVNKIVHVVAFVSNKGLEDRPIAEILACRNLHYLLVYLFICILGLLVHPM 2597
Query 176 CYSVL 180
Y +L
Sbjct 2598 IYCIL 2602
Lambda K H
0.333 0.145 0.472
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2914594326
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40