bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0405_orf2
Length=83
Score E
Sequences producing significant alignments: (Bits) Value
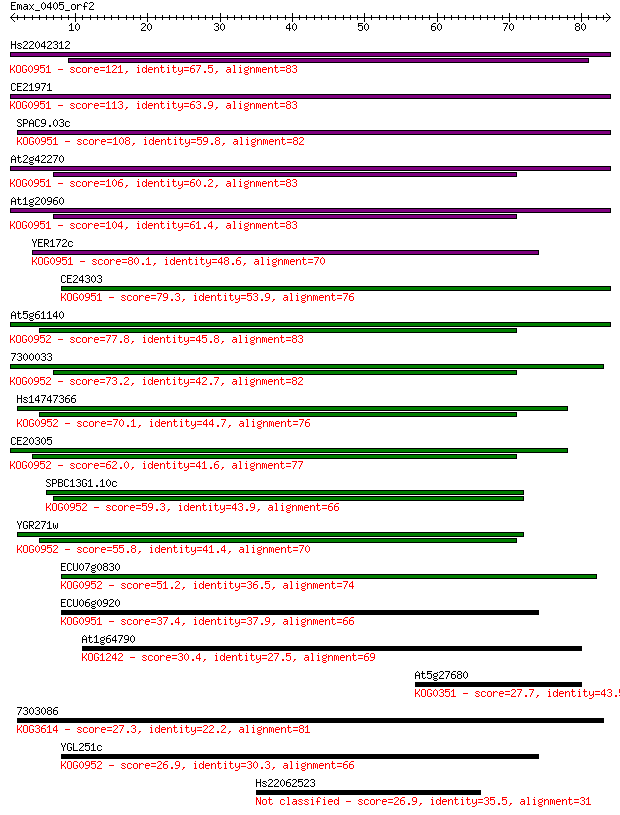
Hs22042312 121 3e-28
CE21971 113 1e-25
SPAC9.03c 108 3e-24
At2g42270 106 8e-24
At1g20960 104 4e-23
YER172c 80.1 8e-16
CE24303 79.3 2e-15
At5g61140 77.8 5e-15
7300033 73.2 1e-13
Hs14747366 70.1 9e-13
CE20305 62.0 3e-10
SPBC13G1.10c 59.3 2e-09
YGR271w 55.8 2e-08
ECU07g0830 51.2 5e-07
ECU06g0920 37.4 0.006
At1g64790 30.4 0.95
At5g27680 27.7 5.6
7303086 27.3 6.6
YGL251c 26.9 8.9
Hs22062523 26.9 9.1
> Hs22042312
Length=2136
Score = 121 bits (304), Expect = 3e-28, Method: Composition-based stats.
Identities = 56/83 (67%), Positives = 68/83 (81%), Gaps = 0/83 (0%)
Query 1 RVKPEKGLFVFGNHYRPVPLKQTYVGIKDKKAIRRYNTMNDVTYEKVLECAGKNQVLIFV 60
RV P KGLF F N +RPVPL+QTYVGI +KKAI+R+ MN++ YEK++E AGKNQVL+FV
Sbjct 666 RVDPAKGLFYFDNSFRPVPLEQTYVGITEKKAIKRFQIMNEIVYEKIMEHAGKNQVLVFV 725
Query 61 HSRKETVKTAKFICDAALQKDTL 83
HSRKET KTA+ I D L+KDTL
Sbjct 726 HSRKETGKTARAIRDMCLEKDTL 748
Score = 35.4 bits (80), Expect = 0.025, Method: Composition-based stats.
Identities = 25/75 (33%), Positives = 36/75 (48%), Gaps = 3/75 (4%)
Query 9 FVFGNHYRPVPLKQTYVGIKDKKAIRRYNTMNDVTYEKVLECAGKNQVLIFVHSRKETVK 68
F F + RPVPL+ G R +M Y + + + K V++FV SRK+T
Sbjct 1512 FNFHPNVRPVPLELHIQGFNISHTQTRLLSMAKPVYHAITKHSPKKPVIVFVPSRKQTRL 1571
Query 69 TAKFI---CDAALQK 80
TA I C A +Q+
Sbjct 1572 TAIDILTTCAADIQR 1586
> CE21971
Length=2145
Score = 113 bits (282), Expect = 1e-25, Method: Composition-based stats.
Identities = 53/83 (63%), Positives = 67/83 (80%), Gaps = 1/83 (1%)
Query 1 RVKPEKGLFVFGNHYRPVPLKQTYVGIKDKKAIRRYNTMNDVTYEKVLECAGKNQVLIFV 60
RVKPE L F N YRPVPL+Q Y+G+ +KKA++R+ MN+V Y+K++E AGK+QVL+FV
Sbjct 660 RVKPEH-LHFFDNSYRPVPLEQQYIGVTEKKALKRFQAMNEVVYDKIMEHAGKSQVLVFV 718
Query 61 HSRKETVKTAKFICDAALQKDTL 83
HSRKET KTAK I DA L+KDTL
Sbjct 719 HSRKETAKTAKAIRDACLEKDTL 741
> SPAC9.03c
Length=2176
Score = 108 bits (269), Expect = 3e-24, Method: Composition-based stats.
Identities = 49/82 (59%), Positives = 63/82 (76%), Gaps = 0/82 (0%)
Query 2 VKPEKGLFVFGNHYRPVPLKQTYVGIKDKKAIRRYNTMNDVTYEKVLECAGKNQVLIFVH 61
V P+KGLF F + YRP PLKQ ++GI +K +R T N+ YEKV++ AGKNQVLIFVH
Sbjct 706 VDPKKGLFYFDSTYRPCPLKQEFIGITEKTPFKRMQTTNEACYEKVMQHAGKNQVLIFVH 765
Query 62 SRKETVKTAKFICDAALQKDTL 83
SRKET KTA+FI D AL+++T+
Sbjct 766 SRKETAKTARFIRDKALEEETI 787
> At2g42270
Length=2172
Score = 106 bits (265), Expect = 8e-24, Method: Composition-based stats.
Identities = 50/83 (60%), Positives = 61/83 (73%), Gaps = 0/83 (0%)
Query 1 RVKPEKGLFVFGNHYRPVPLKQTYVGIKDKKAIRRYNTMNDVTYEKVLECAGKNQVLIFV 60
RV + GLF+F YRPVPL Q Y+GI KK +RR+ MND+ Y+KV+ AGK+QVLIFV
Sbjct 691 RVDLKNGLFIFDRSYRPVPLGQQYIGINVKKPLRRFQLMNDICYQKVVAVAGKHQVLIFV 750
Query 61 HSRKETVKTAKFICDAALQKDTL 83
HSRKET KTA+ I D A+ DTL
Sbjct 751 HSRKETAKTARAIRDTAMANDTL 773
Score = 31.6 bits (70), Expect = 0.37, Method: Composition-based stats.
Identities = 21/65 (32%), Positives = 31/65 (47%), Gaps = 1/65 (1%)
Query 7 GLFVFGNHYRPVPLKQTYVGIKDKKAIRRYNTMNDVTYEKVLECA-GKNQVLIFVHSRKE 65
G+F F + RPVPL+ G+ R M TY +++ A K ++FV +RK
Sbjct 1536 GVFNFPPNVRPVPLEIHIHGVDILSFEARMQAMTKPTYTAIVQHAKNKKPAIVFVPTRKH 1595
Query 66 TVKTA 70
TA
Sbjct 1596 VRLTA 1600
> At1g20960
Length=2171
Score = 104 bits (259), Expect = 4e-23, Method: Composition-based stats.
Identities = 51/83 (61%), Positives = 61/83 (73%), Gaps = 0/83 (0%)
Query 1 RVKPEKGLFVFGNHYRPVPLKQTYVGIKDKKAIRRYNTMNDVTYEKVLECAGKNQVLIFV 60
RV +KGLF F YRPVPL Q Y+GI KK ++R+ MND+ Y+KVL AGK+QVLIFV
Sbjct 690 RVDLKKGLFKFDRSYRPVPLHQQYIGISVKKPLQRFQLMNDLCYQKVLAGAGKHQVLIFV 749
Query 61 HSRKETVKTAKFICDAALQKDTL 83
HSRKET KTA+ I D A+ DTL
Sbjct 750 HSRKETSKTARAIRDTAMANDTL 772
Score = 33.1 bits (74), Expect = 0.13, Method: Composition-based stats.
Identities = 22/65 (33%), Positives = 30/65 (46%), Gaps = 1/65 (1%)
Query 7 GLFVFGNHYRPVPLKQTYVGIKDKKAIRRYNTMNDVTYEKVLECA-GKNQVLIFVHSRKE 65
GLF F RPVPL+ G+ R M TY +++ A K ++FV +RK
Sbjct 1535 GLFNFPPGVRPVPLEIHIQGVDISSFEARMQAMTKPTYTAIVQHAKNKKPAIVFVPTRKH 1594
Query 66 TVKTA 70
TA
Sbjct 1595 VRLTA 1599
> YER172c
Length=2163
Score = 80.1 bits (196), Expect = 8e-16, Method: Composition-based stats.
Identities = 34/71 (47%), Positives = 52/71 (73%), Gaps = 1/71 (1%)
Query 4 PEKGLFVFGNHYRPVPLKQTYVGIKDKKAIRRYNTMNDVTYEKVLECAGK-NQVLIFVHS 62
P++GLF F + +RP PL Q + GIK++ ++++ MND YEKVLE + NQ+++FVHS
Sbjct 687 PKEGLFYFDSSFRPCPLSQQFCGIKERNSLKKLKAMNDACYEKVLESINEGNQIIVFVHS 746
Query 63 RKETVKTAKFI 73
RKET +TA ++
Sbjct 747 RKETSRTATWL 757
> CE24303
Length=1002
Score = 79.3 bits (194), Expect = 2e-15, Method: Composition-based stats.
Identities = 41/77 (53%), Positives = 54/77 (70%), Gaps = 1/77 (1%)
Query 8 LFVFGNHYRPVPLKQTYVGIKDKKAIRRYN-TMNDVTYEKVLECAGKNQVLIFVHSRKET 66
L F N YRPVPLKQ +G+ +K + MN+ Y+K+++ AGK+QVL+FVH RKET
Sbjct 443 LHFFDNSYRPVPLKQQIIGVTEKNDDSATSQAMNEAVYDKIMKHAGKSQVLVFVHYRKET 502
Query 67 VKTAKFICDAALQKDTL 83
+TAK I DA L+KDTL
Sbjct 503 TETAKAIRDACLEKDTL 519
> At5g61140
Length=2137
Score = 77.8 bits (190), Expect = 5e-15, Method: Composition-based stats.
Identities = 38/84 (45%), Positives = 55/84 (65%), Gaps = 1/84 (1%)
Query 1 RVKPEKGLFVFGNHYRPVPLKQTYVGIKDKKAIRRYNTMNDVTYEKVLECAGK-NQVLIF 59
RV + GLF F + YRPVPL Q Y+GI + R +N++ Y+KV++ + +Q +IF
Sbjct 692 RVNTDTGLFYFDSSYRPVPLAQQYIGITEHNFAARNELLNEICYKKVVDSIKQGHQAMIF 751
Query 60 VHSRKETVKTAKFICDAALQKDTL 83
VHSRK+T KTA+ + D A Q +TL
Sbjct 752 VHSRKDTSKTAEKLVDLARQYETL 775
Score = 43.9 bits (102), Expect = 7e-05, Method: Composition-based stats.
Identities = 26/66 (39%), Positives = 33/66 (50%), Gaps = 0/66 (0%)
Query 5 EKGLFVFGNHYRPVPLKQTYVGIKDKKAIRRYNTMNDVTYEKVLECAGKNQVLIFVHSRK 64
E GLF F RPVP++ G K R N+MN Y + + VLIFV SR+
Sbjct 1522 EIGLFNFKPSVRPVPIEVHIQGYPGKYYCPRMNSMNKPAYAAICTHSPTKPVLIFVSSRR 1581
Query 65 ETVKTA 70
+T TA
Sbjct 1582 QTRLTA 1587
> 7300033
Length=1809
Score = 73.2 bits (178), Expect = 1e-13, Method: Composition-based stats.
Identities = 35/83 (42%), Positives = 53/83 (63%), Gaps = 1/83 (1%)
Query 1 RVKPEKGLFVFGNHYRPVPLKQTYVGIKDKKAIRRYNTMNDVTYEKVLECAGK-NQVLIF 59
RV P KGLF F + +RPVPL +VGIK K +++ M+ Y+K +E + +Q+++F
Sbjct 298 RVNPMKGLFYFDSRFRPVPLDTNFVGIKSVKQLQQIADMDQCCYQKCVEMVQEGHQIMVF 357
Query 60 VHSRKETVKTAKFICDAALQKDT 82
VH+R TV+TA I + A Q +T
Sbjct 358 VHARNATVRTANVIRELAQQNNT 380
Score = 38.5 bits (88), Expect = 0.003, Method: Composition-based stats.
Identities = 22/64 (34%), Positives = 32/64 (50%), Gaps = 0/64 (0%)
Query 7 GLFVFGNHYRPVPLKQTYVGIKDKKAIRRYNTMNDVTYEKVLECAGKNQVLIFVHSRKET 66
GL+ F RPVPL+ G K R TMN T++ + + ++FV SR++T
Sbjct 1146 GLYNFKPSVRPVPLQVHINGFPGKHYCPRMATMNRPTFQAIRTYSPCEPTIVFVSSRRQT 1205
Query 67 VKTA 70
TA
Sbjct 1206 RLTA 1209
> Hs14747366
Length=1866
Score = 70.1 bits (170), Expect = 9e-13, Method: Composition-based stats.
Identities = 34/77 (44%), Positives = 50/77 (64%), Gaps = 1/77 (1%)
Query 2 VKPEKGLFVFGNHYRPVPLKQTYVGIKDKKAIRRYNTMNDVTYEKVL-ECAGKNQVLIFV 60
V P GLF F +RPVPL QT++GIK +++ N M++V YE VL + +QV++FV
Sbjct 327 VNPYIGLFFFDGRFRPVPLGQTFLGIKCANKMQQLNNMDEVCYENVLKQVKAGHQVMVFV 386
Query 61 HSRKETVKTAKFICDAA 77
H+R TV+TA + + A
Sbjct 387 HARNATVRTAMSLIERA 403
Score = 39.7 bits (91), Expect = 0.001, Method: Composition-based stats.
Identities = 23/66 (34%), Positives = 33/66 (50%), Gaps = 0/66 (0%)
Query 5 EKGLFVFGNHYRPVPLKQTYVGIKDKKAIRRYNTMNDVTYEKVLECAGKNQVLIFVHSRK 64
+ GLF F RPVPL+ G + R +MN ++ + + VLIFV SR+
Sbjct 1171 QMGLFNFRPSVRPVPLEVHIQGFPGQHYCPRMASMNKPAFQAIRSHSPAKPVLIFVSSRR 1230
Query 65 ETVKTA 70
+T TA
Sbjct 1231 QTRLTA 1236
> CE20305
Length=1798
Score = 62.0 bits (149), Expect = 3e-10, Method: Composition-based stats.
Identities = 32/79 (40%), Positives = 47/79 (59%), Gaps = 2/79 (2%)
Query 1 RVKPEKGLFVFGNHYRPVPLKQTYVGIKDKKAIRRYNT-MNDVTYEKVLECAGK-NQVLI 58
RV P KGLF F +RPVPL Q ++G + R T M++V Y++V++ + +QVL+
Sbjct 279 RVNPYKGLFFFDGRFRPVPLTQKFIGTRKAGNFRDNLTLMDNVCYDEVVDFVKRGHQVLV 338
Query 59 FVHSRKETVKTAKFICDAA 77
FVH+R T K + C A
Sbjct 339 FVHTRNGTAKLGEAFCARA 357
Score = 40.4 bits (93), Expect = 7e-04, Method: Composition-based stats.
Identities = 22/67 (32%), Positives = 33/67 (49%), Gaps = 0/67 (0%)
Query 4 PEKGLFVFGNHYRPVPLKQTYVGIKDKKAIRRYNTMNDVTYEKVLECAGKNQVLIFVHSR 63
P++ + F RPVP+ G + R MN Y+ +L + + VLIFV SR
Sbjct 1143 PDEACYNFRPSVRPVPISVHIQGFPGQHYCPRMGLMNKPAYKAILTYSPRKPVLIFVSSR 1202
Query 64 KETVKTA 70
++T TA
Sbjct 1203 RQTRLTA 1209
> SPBC13G1.10c
Length=1935
Score = 59.3 bits (142), Expect = 2e-09, Method: Composition-based stats.
Identities = 29/67 (43%), Positives = 46/67 (68%), Gaps = 2/67 (2%)
Query 6 KGLFVFGNHYRPVPLKQTYVGIKDKKAIRRYNTMNDVTYEKVLECAGK-NQVLIFVHSRK 64
KGLF F + +RP P++Q ++G K I N +++ ++KVL+ + +QV+IFVHSRK
Sbjct 477 KGLFYFSSAFRPCPIEQHFIGAKGSPKIVNSN-IDEACFDKVLKLIQEGHQVMIFVHSRK 535
Query 65 ETVKTAK 71
ET+ +AK
Sbjct 536 ETINSAK 542
Score = 42.0 bits (97), Expect = 3e-04, Method: Composition-based stats.
Identities = 24/65 (36%), Positives = 34/65 (52%), Gaps = 0/65 (0%)
Query 7 GLFVFGNHYRPVPLKQTYVGIKDKKAIRRYNTMNDVTYEKVLECAGKNQVLIFVHSRKET 66
GLF F + RPVPL+ G + R +MN ++ + + VLIFV SR++T
Sbjct 1316 GLFNFRHSVRPVPLEIYIDGFPGRAYCPRMMSMNKPAFQAIKTHSPTQPVLIFVSSRRQT 1375
Query 67 VKTAK 71
TAK
Sbjct 1376 RLTAK 1380
> YGR271w
Length=1967
Score = 55.8 bits (133), Expect = 2e-08, Method: Composition-based stats.
Identities = 29/72 (40%), Positives = 45/72 (62%), Gaps = 2/72 (2%)
Query 2 VKPEKGLFVFGNHYRPVPLKQTYVGIKDKKAIRRYN-TMNDVTYEKVLECAGKN-QVLIF 59
V + G+F F +RP PL+Q +G + K R+ ++ V Y+K+ E + QV++F
Sbjct 479 VNRQIGMFYFDQSFRPKPLEQQLLGCRGKAGSRQSKENIDKVAYDKLSEMIQRGYQVMVF 538
Query 60 VHSRKETVKTAK 71
VHSRKETVK+A+
Sbjct 539 VHSRKETVKSAR 550
Score = 37.0 bits (84), Expect = 0.008, Method: Composition-based stats.
Identities = 24/67 (35%), Positives = 33/67 (49%), Gaps = 1/67 (1%)
Query 5 EKGLFVFGNHYRPVPLKQTYVGIKDKKAI-RRYNTMNDVTYEKVLECAGKNQVLIFVHSR 63
+ GL+ F + RPVPLK G D A TMN + + + + LIFV SR
Sbjct 1320 DHGLYNFPSSVRPVPLKMYIDGFPDNLAFCPLMKTMNKPVFMAIKQHSPDKPALIFVASR 1379
Query 64 KETVKTA 70
++T TA
Sbjct 1380 RQTRLTA 1386
> ECU07g0830
Length=1058
Score = 51.2 bits (121), Expect = 5e-07, Method: Composition-based stats.
Identities = 27/75 (36%), Positives = 45/75 (60%), Gaps = 4/75 (5%)
Query 8 LFVFGNHYRPVPLKQTYVGIKDKKAIRRYNTMNDVTYEKVLE-CAGKNQVLIFVHSRKET 66
+F F YRPVPLK + +G+K K + +++ EKV E + QVL+FVHSR +T
Sbjct 281 VFSFDQGYRPVPLKMSVIGMKRKS---KPQLEDELLQEKVREYLSNGKQVLVFVHSRGDT 337
Query 67 VKTAKFICDAALQKD 81
++ A+ + D +++
Sbjct 338 IRIARLLGDEGERRN 352
> ECU06g0920
Length=1481
Score = 37.4 bits (85), Expect = 0.006, Method: Composition-based stats.
Identities = 25/69 (36%), Positives = 37/69 (53%), Gaps = 11/69 (15%)
Query 8 LFVFGNHYR--PVPLKQTYVGIKDKKAIRRYNTMNDVTYEKVLECAGKN-QVLIFVHSRK 64
+F FG +R P+ + VG ++++ D+T EKVLE N VL+FVHSR
Sbjct 419 IFYFGPEFRKSPIDYEVINVGAREREM--------DMTIEKVLENLDSNGPVLVFVHSRN 470
Query 65 ETVKTAKFI 73
E ++ A I
Sbjct 471 EALEVANEI 479
> At1g64790
Length=2428
Score = 30.4 bits (67), Expect = 0.95, Method: Composition-based stats.
Identities = 19/69 (27%), Positives = 35/69 (50%), Gaps = 4/69 (5%)
Query 11 FGNHYRPVPLKQTYVGIKDKKAIRRYNTMNDVTYEKVLECAGKNQVLIFVHSRKETVKTA 70
G P+ + G+KD +R + +V+ AG++Q+L F+ T++TA
Sbjct 1716 LGERVLPLIIPILSKGLKDPDVDKRQGVC--IGLNEVMASAGRSQLLSFMDQLIPTIRTA 1773
Query 71 KFICDAALQ 79
+CD+AL+
Sbjct 1774 --LCDSALE 1780
> At5g27680
Length=819
Score = 27.7 bits (60), Expect = 5.6, Method: Composition-based stats.
Identities = 10/23 (43%), Positives = 17/23 (73%), Gaps = 0/23 (0%)
Query 57 LIFVHSRKETVKTAKFICDAALQ 79
+I+V +RKE+V AK++C L+
Sbjct 488 IIYVPTRKESVNIAKYLCGVGLK 510
> 7303086
Length=1998
Score = 27.3 bits (59), Expect = 6.6, Method: Composition-based stats.
Identities = 18/82 (21%), Positives = 37/82 (45%), Gaps = 6/82 (7%)
Query 2 VKPEKGLFVFGNHYRPVPLKQTYVGIKDKKAIRRYNTMNDVTYEKV-LECAGKNQVLIFV 60
V+P+ +F G H+ VPL + + + ++ +N+ K ++ + + I V
Sbjct 1372 VRPDAYIFDEGTHFEVVPLPE-----EPDEVVKSREALNEQVVRKASMQSEADSDIYIPV 1426
Query 61 HSRKETVKTAKFICDAALQKDT 82
R T +T K +++DT
Sbjct 1427 SQRPSTCETVKRTPYVTVRQDT 1448
> YGL251c
Length=1187
Score = 26.9 bits (58), Expect = 8.9, Method: Composition-based stats.
Identities = 20/68 (29%), Positives = 29/68 (42%), Gaps = 2/68 (2%)
Query 8 LFVFGNHYRPVPLKQTYVG--IKDKKAIRRYNTMNDVTYEKVLECAGKNQVLIFVHSRKE 65
+ F YR V L + G K ++ N E + + A VLIF +R
Sbjct 324 ILSFDESYRQVQLTKFVYGYSFNCKNDFQKDAIYNSKLIEIIEKHADNRPVLIFCPTRAS 383
Query 66 TVKTAKFI 73
T+ TAKF+
Sbjct 384 TISTAKFL 391
> Hs22062523
Length=442
Score = 26.9 bits (58), Expect = 9.1, Method: Composition-based stats.
Identities = 11/31 (35%), Positives = 17/31 (54%), Gaps = 0/31 (0%)
Query 35 RYNTMNDVTYEKVLECAGKNQVLIFVHSRKE 65
R ++N+ EKV+E G +V+ F H E
Sbjct 361 RMGSINEKNIEKVVEVTGDERVMAFTHDEME 391
Lambda K H
0.322 0.137 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1200681374
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40