bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0413_orf2
Length=156
Score E
Sequences producing significant alignments: (Bits) Value
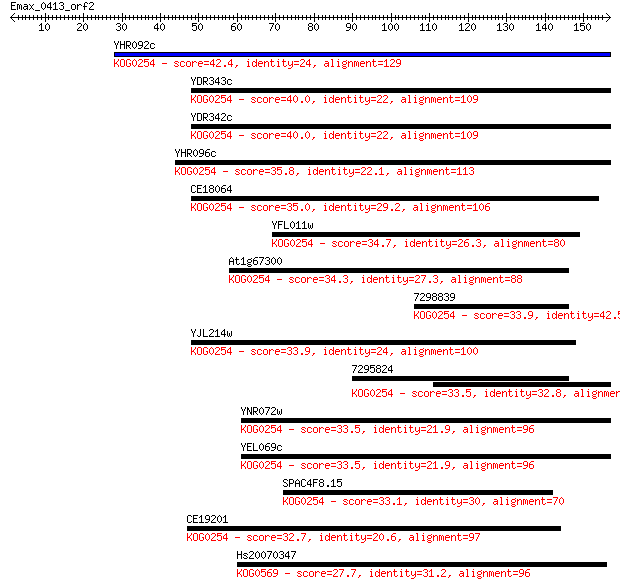
YHR092c 42.4 3e-04
YDR343c 40.0 0.002
YDR342c 40.0 0.002
YHR096c 35.8 0.034
CE18064 35.0 0.061
YFL011w 34.7 0.074
At1g67300 34.3 0.11
7298839 33.9 0.12
YJL214w 33.9 0.13
7295824 33.5 0.17
YNR072w 33.5 0.19
YEL069c 33.5 0.19
SPAC4F8.15 33.1 0.24
CE19201 32.7 0.27
Hs20070347 27.7 8.6
> YHR092c
Length=560
Score = 42.4 bits (98), Expect = 3e-04, Method: Composition-based stats.
Identities = 31/129 (24%), Positives = 53/129 (41%), Gaps = 3/129 (2%)
Query 28 SAPVPAAAATAVATKPSAGAAETAAAAAAAARILQQKKRRKIIISLLLAGALGCMHSLLA 87
SA PA A + + AE A A+ I K K+ L++ + + L
Sbjct 289 SADDPAVMAEVEVVQATV-EAEKLAGNASWGEIFSTKT--KVFQRLIMGAMIQSLQQLTG 345
Query 88 ANTLIVFISDVLLILGVCDNKGPAMVIGFAKLAGALSCLLFADSVGRRVLLLWGAGGIFA 147
N + + V +G+ D+ ++V+G A + + GRR LLWGA + A
Sbjct 346 DNYFFYYGTTVFTAVGLEDSFETSIVLGIVNFASTFVGIFLVERYGRRRCLLWGAASMTA 405
Query 148 THMGLSASG 156
+ ++ G
Sbjct 406 CMVVFASVG 414
> YDR343c
Length=570
Score = 40.0 bits (92), Expect = 0.002, Method: Composition-based stats.
Identities = 24/109 (22%), Positives = 47/109 (43%), Gaps = 2/109 (1%)
Query 48 AETAAAAAAAARILQQKKRRKIIISLLLAGALGCMHSLLAANTLIVFISDVLLILGVCDN 107
AE A A+ + K K++ L++ + + L N + + + +G+ D+
Sbjct 302 AEKLAGNASWGELFSSKT--KVLQRLIMGAMIQSLQQLTGDNYFFYYGTTIFKAVGLSDS 359
Query 108 KGPAMVIGFAKLAGALSCLLFADSVGRRVLLLWGAGGIFATHMGLSASG 156
++V+G A + + GRR LLWGA + A + ++ G
Sbjct 360 FETSIVLGIVNFASTFVGIYVVERYGRRTCLLWGAASMTACMVVYASVG 408
> YDR342c
Length=570
Score = 40.0 bits (92), Expect = 0.002, Method: Composition-based stats.
Identities = 24/109 (22%), Positives = 47/109 (43%), Gaps = 2/109 (1%)
Query 48 AETAAAAAAAARILQQKKRRKIIISLLLAGALGCMHSLLAANTLIVFISDVLLILGVCDN 107
AE A A+ + K K++ L++ + + L N + + + +G+ D+
Sbjct 302 AEKLAGNASWGELFSSKT--KVLQRLIMGAMIQSLQQLTGDNYFFYYGTTIFKAVGLSDS 359
Query 108 KGPAMVIGFAKLAGALSCLLFADSVGRRVLLLWGAGGIFATHMGLSASG 156
++V+G A + + GRR LLWGA + A + ++ G
Sbjct 360 FETSIVLGIVNFASTFVGIYVVERYGRRTCLLWGAASMTACMVVYASVG 408
> YHR096c
Length=592
Score = 35.8 bits (81), Expect = 0.034, Method: Composition-based stats.
Identities = 25/115 (21%), Positives = 51/115 (44%), Gaps = 6/115 (5%)
Query 44 SAGAAETAAAAAAAARILQQKKR--RKIIISLLLAGALGCMHSLLAANTLIVFISDVLLI 101
S+ AE A +A+ ++ K + R+ ++ +++ + L N + + +
Sbjct 319 SSIEAERLAGSASWGELVTGKPQMFRRTLMGMMIQS----LQQLTGDNYFFYYGTTIFQA 374
Query 102 LGVCDNKGPAMVIGFAKLAGALSCLLFADSVGRRVLLLWGAGGIFATHMGLSASG 156
+G+ D+ A+V+G L D GRR LLWG G+ ++ ++ G
Sbjct 375 VGLEDSFETAIVLGVVNFVSTFFSLYTVDRFGRRNCLLWGCVGMICCYVVYASVG 429
> CE18064
Length=639
Score = 35.0 bits (79), Expect = 0.061, Method: Composition-based stats.
Identities = 31/111 (27%), Positives = 50/111 (45%), Gaps = 8/111 (7%)
Query 48 AETAAAAAAAARILQQKKRRKIIISLLLAGALGCMHSLLAANTLIVFISDVLLILGVCDN 107
AE A RIL+ RK ++++ L L NT++ + +++ GV DN
Sbjct 296 AEHAGNGPTIIRILKTPHVRK---AMIIGSLLQMFQQLSGINTVMYYTGNIIRSAGVKDN 352
Query 108 KGPAMV-IGFAKL--AGALSCLLFADSVGRRVLLLWGAGGI--FATHMGLS 153
+ +G + + G + + VGRRVLLL G+ F MG+S
Sbjct 353 HTTIWISVGTSAINFLGTFIPIALVERVGRRVLLLVSMIGVILFLIAMGVS 403
> YFL011w
Length=546
Score = 34.7 bits (78), Expect = 0.074, Method: Composition-based stats.
Identities = 21/86 (24%), Positives = 37/86 (43%), Gaps = 6/86 (6%)
Query 69 IIISLLLAGALGCMHSLLAANTLIVFISDVLLILGVCDNKGPAMVIGFAKLAGALSCLLF 128
I+ +++ + + L N + + + +G+ D+ ++V+G A L
Sbjct 305 ILPRVIMGIVIQSLQQLTGCNYFFYYGTTIFNAVGMQDSFETSIVLGAVNFASTFVALYI 364
Query 129 ADSVGRRVLLLWGAGG------IFAT 148
D GRR LLWG+ IFAT
Sbjct 365 VDKFGRRKCLLWGSASMAICFVIFAT 390
> At1g67300
Length=493
Score = 34.3 bits (77), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 24/88 (27%), Positives = 41/88 (46%), Gaps = 5/88 (5%)
Query 58 ARILQQKKRRKIIISLLLAGALGCMHSLLAANTLIVFISDVLLILGVCDNKGPAMVIGFA 117
+ +L + R + I L + L N + F S V GV + G + +G +
Sbjct 283 SELLYGRHSRVVFI----GSTLFALQQLSGINAVFYFSSTVFKSAGVPSDLG-NIFVGVS 337
Query 118 KLAGALSCLLFADSVGRRVLLLWGAGGI 145
L G++ ++ D VGR++LLLW G+
Sbjct 338 NLLGSVIAMVLMDKVGRKLLLLWSFIGM 365
> 7298839
Length=438
Score = 33.9 bits (76), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 17/40 (42%), Positives = 24/40 (60%), Gaps = 0/40 (0%)
Query 106 DNKGPAMVIGFAKLAGALSCLLFADSVGRRVLLLWGAGGI 145
D +V+G A+L G S ++ D +GRRVLLL GG+
Sbjct 278 DPNTSTIVVGAAQLVGIFSAVVLVDRLGRRVLLLTSMGGM 317
> YJL214w
Length=569
Score = 33.9 bits (76), Expect = 0.13, Method: Composition-based stats.
Identities = 24/100 (24%), Positives = 42/100 (42%), Gaps = 2/100 (2%)
Query 48 AETAAAAAAAARILQQKKRRKIIISLLLAGALGCMHSLLAANTLIVFISDVLLILGVCDN 107
AE A A+ + +K K+ L + + + L N + + + +G+ D+
Sbjct 304 AEEAMGTASWKELFSRKT--KVFQRLTMTVMINSLQQLTGDNYFFYYGTTIFKSVGMNDS 361
Query 108 KGPAMVIGFAKLAGALSCLLFADSVGRRVLLLWGAGGIFA 147
++V+G A L D +GRR LL GA + A
Sbjct 362 FETSIVLGIVNFASCFFSLYSVDKLGRRRCLLLGAATMTA 401
> 7295824
Length=780
Score = 33.5 bits (75), Expect = 0.17, Method: Composition-based stats.
Identities = 18/57 (31%), Positives = 30/57 (52%), Gaps = 1/57 (1%)
Query 90 TLIVFISDVLLILG-VCDNKGPAMVIGFAKLAGALSCLLFADSVGRRVLLLWGAGGI 145
+ I ++SD+ G V D ++IG ++ G + + D VGRRVL+L G+
Sbjct 602 SFINYMSDIFKASGSVVDVNTATIIIGLVQIVGVYTSTILVDIVGRRVLMLISTMGV 658
Score = 28.9 bits (63), Expect = 4.3, Method: Composition-based stats.
Identities = 16/46 (34%), Positives = 27/46 (58%), Gaps = 3/46 (6%)
Query 111 AMVIGFAKLAGALSCLLFADSVGRRVLLLWGAGGIFATHMGLSASG 156
A+++G +L G + + + +GR++LLL A GI +G SA G
Sbjct 237 AIIVGVIQLMGTYASTVLVERLGRKILLLVSAVGI---GLGQSAMG 279
> YNR072w
Length=564
Score = 33.5 bits (75), Expect = 0.19, Method: Composition-based stats.
Identities = 21/96 (21%), Positives = 45/96 (46%), Gaps = 4/96 (4%)
Query 61 LQQKKRRKIIISLLLAGALGCMHSLLAANTLIVFISDVLLILGVCDNKGPAMVIGFAKLA 120
++ K +++I +L+ L L N + + + +G+ D ++V+G
Sbjct 310 VKTKVLQRLITGILVQTFL----QLTGENYFFFYGTTIFKSVGLTDGFETSIVLGTVNFF 365
Query 121 GALSCLLFADSVGRRVLLLWGAGGIFATHMGLSASG 156
+ ++ D +GRR LL+GA G+ A + ++ G
Sbjct 366 STIIAVMVVDKIGRRKCLLFGAAGMMACMVIFASIG 401
> YEL069c
Length=564
Score = 33.5 bits (75), Expect = 0.19, Method: Composition-based stats.
Identities = 21/96 (21%), Positives = 45/96 (46%), Gaps = 4/96 (4%)
Query 61 LQQKKRRKIIISLLLAGALGCMHSLLAANTLIVFISDVLLILGVCDNKGPAMVIGFAKLA 120
++ K +++I +L+ L L N + + + +G+ D ++V+G
Sbjct 310 VKTKVLQRLITGILVQTFL----QLTGENYFFFYGTTIFKSVGLTDGFETSIVLGTVNFF 365
Query 121 GALSCLLFADSVGRRVLLLWGAGGIFATHMGLSASG 156
+ ++ D +GRR LL+GA G+ A + ++ G
Sbjct 366 STIIAVMVVDKIGRRKCLLFGAAGMMACMVIFASIG 401
> SPAC4F8.15
Length=286
Score = 33.1 bits (74), Expect = 0.24, Method: Compositional matrix adjust.
Identities = 21/73 (28%), Positives = 33/73 (45%), Gaps = 3/73 (4%)
Query 72 SLLLAGALGCMHSLLAANTLIVFISDVLLILGVCDNKGPAMVIGFAKLAGALSCLLFADS 131
+L+LA L M L N+L+ F S + ++G + ++I + D
Sbjct 49 ALILACGLQAMQQLSGFNSLMYFSSTIFEVVGFNNPTATGLIIAATNFVFTIVAFGVIDF 108
Query 132 VGRRVLLL---WG 141
GRR+LLL WG
Sbjct 109 FGRRILLLLTVWG 121
> CE19201
Length=606
Score = 32.7 bits (73), Expect = 0.27, Method: Composition-based stats.
Identities = 20/100 (20%), Positives = 43/100 (43%), Gaps = 3/100 (3%)
Query 47 AAETAAAAAAAARILQQKKRRKIIISLLLAGALGCMHSLLAANTLIVFISDVLLILGVCD 106
A E A A+ I + K ++ + + L L NT++ + +D++ G+ +
Sbjct 254 AKENEKAHASGPVIWRILKTPHVLKACFIGSMLQAFQQLAGINTILYYTADIIRSSGISN 313
Query 107 NKGP---AMVIGFAKLAGALSCLLFADSVGRRVLLLWGAG 143
N ++++ G + + VGRR++ L+ G
Sbjct 314 NHTTIWISVLLSLCNFIGPFVPMSLIEKVGRRIIFLFSCG 353
> Hs20070347
Length=503
Score = 27.7 bits (60), Expect = 8.6, Method: Compositional matrix adjust.
Identities = 30/105 (28%), Positives = 49/105 (46%), Gaps = 13/105 (12%)
Query 60 ILQQKKRRKIIISLLLAGALGCMHSLLAANTLIVFISDVLLILGVCDNKGPAMVIGFAK- 118
+ Q + R+ + SL++ LG L +++ + S V GV + K +IG
Sbjct 271 LFQHRALRRQVTSLVV---LGSAMELCGNDSVYAYASSVFRKAGVPEAKIQYAIIGTGSC 327
Query 119 --LAGALSCLLFADSVGRRVLLLWGA------GGIFATHMGLSAS 155
L +SC++ + VGRRVLL+ G G IF + L +S
Sbjct 328 ELLTAVVSCVVI-ERVGRRVLLIGGYSLMTCWGSIFTVALCLQSS 371
Lambda K H
0.321 0.131 0.370
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2070320142
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40