bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0442_orf1
Length=249
Score E
Sequences producing significant alignments: (Bits) Value
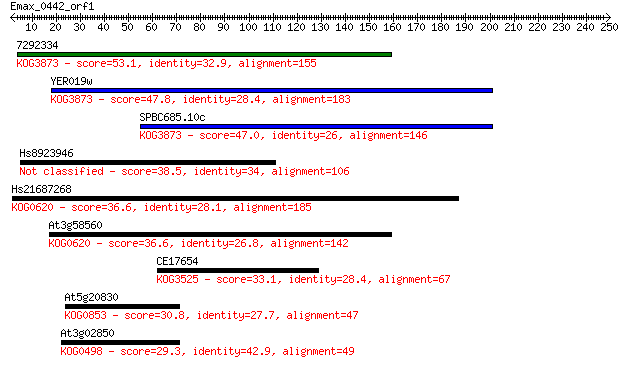
7292334 53.1 5e-07
YER019w 47.8 2e-05
SPBC685.10c 47.0 4e-05
Hs8923946 38.5 0.012
Hs21687268 36.6 0.050
At3g58560 36.6 0.053
CE17654 33.1 0.62
At5g20830 30.8 2.7
At3g02850 29.3 8.7
> 7292334
Length=402
Score = 53.1 bits (126), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 51/166 (30%), Positives = 76/166 (45%), Gaps = 22/166 (13%)
Query 4 RLHHIPAALLSTGADIICLQEVYGDNHADFIVESTRHRLPFVGRRTSGGRFSLHNGLMVL 63
R+ I L S DI+ LQEV+ ++ + + T LP SG + GL+VL
Sbjct 26 RIDAICKELASGKYDIVSLQEVWAQEDSELLQKGTEAVLPHSHYFHSG---VMGAGLLVL 82
Query 64 SKYPIVHTQFHKFEDVTYIERC-----FASKGMLEVAVDVPSIGVIAIFNIHLASGAVDP 118
SKYPI+ T FH + Y R F KG+ + V ++ ++N HL + +
Sbjct 83 SKYPILGTLFHAWSVNGYFHRIQHADWFGGKGVGLCRILVGG-QMVHLYNAHLHAEYDNA 141
Query 119 ESRYVEDVRAAEIRQLLKACDAA----ADRGHVPLVI--GDLNAAP 158
Y + ++++A D A A RG+ L I GDLNA P
Sbjct 142 NDEY-------KTHRVIQAFDTAQFIEATRGNSALQILAGDLNAQP 180
> YER019w
Length=477
Score = 47.8 bits (112), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 52/190 (27%), Positives = 83/190 (43%), Gaps = 13/190 (6%)
Query 18 DIICLQEVYGDNHADFIVESTRHRLPFVGRRTSGGRFSLHNGLMVLSKYPIVHTQFHKFE 77
D+I LQE++ ++ + + P+ SG GL +LSK PI T ++F
Sbjct 94 DVIALQEIWCVEDWKYLASACASKYPYQRLFHSG--ILTGPGLAILSKVPIESTFLYRFP 151
Query 78 DVTYIERCFASKGML--EVAVDVPSIGV--IAIFNIHL-ASGAVDPESRYV--EDVRAAE 130
F + +A+ V + G IAI N H+ A A ++ Y+ +A +
Sbjct 152 INGRPSAVFRGDWYVGKSIAITVLNTGTRPIAIMNSHMHAPYAKQGDAAYLCHRSCQAWD 211
Query 131 IRQLLKACDAAADRGHVPLVIGDLNAAPHLCASNYKCFVERGWRDCWLHVHGHQRHLLQH 190
+L+K A G+ +V+GDLN+ P + E G D W +HG Q +
Sbjct 212 FSRLIKLYRQA---GYAVIVVGDLNSRPGSLPHKFLT-QEAGLVDSWEQLHGKQDLAVIA 267
Query 191 HLLHQQQLLE 200
L QQLL+
Sbjct 268 RLSPLQQLLK 277
> SPBC685.10c
Length=368
Score = 47.0 bits (110), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 38/156 (24%), Positives = 70/156 (44%), Gaps = 16/156 (10%)
Query 55 SLHNGLMVLSKYPIVHTQFHKFE-----DVTYIERCFASKGMLEVAVDVPSIGVIAIFNI 109
++ GL + SK+PI+ + +K+ + + KG+ ++ PS +I++FN
Sbjct 21 AMGAGLAMFSKFPIIESSMNKYPLNGRPQAFWRGDWYVGKGVATASLQHPSGRIISLFNT 80
Query 110 HLASGAVDPESRYVEDVRAAEIRQLLKACDAAADRGHVPLVIGDLN----AAPHLCASNY 165
HL + Y+ R ++ + K AA RGH+ + GD N + PH ++Y
Sbjct 81 HLHAPYGKGADTYLCH-RLSQAWYISKLLRAAVQRGHIVIAAGDFNIQPLSVPHEIITSY 139
Query 166 KCFVERGWRDCWLHVHGHQ-RHLLQHHLLHQQQLLE 200
D WL V+ Q H ++ ++L+E
Sbjct 140 GLV-----NDAWLSVYPDQVEHPPNRFSMNDKELVE 170
> Hs8923946
Length=655
Score = 38.5 bits (88), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 36/124 (29%), Positives = 52/124 (41%), Gaps = 23/124 (18%)
Query 5 LHHIPAALLSTGADIICLQEVYGDNHADFIVESTRHRLPF----VGRRTSGGRFS---LH 57
H +A D +CLQEV+ A + E + VG G S L+
Sbjct 345 FDHEVSAFFPANLDFLCLQEVFDKRAATKLKEQLHGYFEYILYDVGVYGCQGCCSFKCLN 404
Query 58 NGLMVLSKYPIVHTQFHKFEDVTYIERC----FASKGMLEVAVDVPS-------IGVIAI 106
+GL+ S+YPI+ +H Y +C ASKG L + V V S +G IA
Sbjct 405 SGLLFASRYPIMDVAYH-----CYPNKCNDDALASKGALFLKVQVGSTPQDQRIVGYIAC 459
Query 107 FNIH 110
++H
Sbjct 460 THLH 463
> Hs21687268
Length=475
Score = 36.6 bits (83), Expect = 0.050, Method: Compositional matrix adjust.
Identities = 52/243 (21%), Positives = 92/243 (37%), Gaps = 65/243 (26%)
Query 2 QRRLHHIPAALLSTGADIICLQEVYGDNHADFIVESTRHRLPFVGRRTSGGRFSLHNGLM 61
+ R I +++ ADII LQEV + + LP + R G FS +
Sbjct 191 EYRKKGIMEEIVNCDADIISLQEVETEQYFTLF-------LPALKERGYDGFFSPKSRAK 243
Query 62 VLSKYPIVHTQ----FHKFEDVTYIER------------CFASKGMLEVAVDVPSIGVIA 105
++S+ H F K E T +++ S+ ML + +IGV
Sbjct 244 IMSEQERKHVDGCAIFFKTEKFTLVQKHTVEFNQVAMANSDGSEAMLNRVMTKDNIGVAV 303
Query 106 IFNIH---LASGA-------------------VDPESRYVEDVRA----AEIRQLLKACD 139
+ +H +G DPE V+ ++ +E++ +L+
Sbjct 304 VLEVHKELFGAGMKPIHAADKQLLIVANAHMHWDPEYSDVKLIQTMMFVSEVKNILEKAS 363
Query 140 A-----AADRGHVPLVI-GDLNAAPHL----------CASNYKCFVERGWRDCWLHVHGH 183
+ AD +PLV+ DLN+ P A N+K F E G+ +C ++ +
Sbjct 364 SRPGSPTADPNSIPLVLCADLNSLPDSGVVEYLSNGGVADNHKDFKELGYNECLMNFSCN 423
Query 184 QRH 186
++
Sbjct 424 GKN 426
> At3g58560
Length=597
Score = 36.6 bits (83), Expect = 0.053, Method: Compositional matrix adjust.
Identities = 38/172 (22%), Positives = 66/172 (38%), Gaps = 37/172 (21%)
Query 17 ADIICLQEVYGDNHADFIVESTRHRLPFVGRRTSGGRFSLHNGLMVLSKYPIVH--TQFH 74
ADI+CLQEV D+ +F LP + + G F + + + F
Sbjct 292 ADIVCLQEVQNDHFEEFF-------LPELDKHGYQGLFKRKTNEVFIGNTNTIDGCATFF 344
Query 75 KFEDVTYIERCFASKGMLEVAVDVP------------SIGVIAIFNIHLASGAVD-PESR 121
+ + +++E A++ + E + V ++ +I + S A D P R
Sbjct 345 RRDRFSHVEFNKAAQSLTEAIIPVSQKKNALNRLVKDNVALIVVLEAKFGSQAADNPGKR 404
Query 122 Y--------------VEDVRAAEIRQLLKACDAAADRGHVP-LVIGDLNAAP 158
++DV+ ++ LLK + A +P LV GD N P
Sbjct 405 QLLCVANTHVNVPHELKDVKLWQVHTLLKGLEKIAASADIPMLVCGDFNTVP 456
> CE17654
Length=760
Score = 33.1 bits (74), Expect = 0.62, Method: Composition-based stats.
Identities = 19/69 (27%), Positives = 36/69 (52%), Gaps = 2/69 (2%)
Query 62 VLSKYPIVHTQFHKFEDVTYIERCFASKGMLEVAVDVPSIG--VIAIFNIHLASGAVDPE 119
+ K+P T+F KF + IE+ F + L+V ++P+ + +++ GA + +
Sbjct 561 IFQKFPGTSTRFQKFPKILEIEKMFRNSKKLQVFRNIPNGNRLQLQLYSDGCYGGADENK 620
Query 120 SRYVEDVRA 128
YVE V+A
Sbjct 621 VSYVEHVQA 629
> At5g20830
Length=808
Score = 30.8 bits (68), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 13/47 (27%), Positives = 24/47 (51%), Gaps = 2/47 (4%)
Query 24 EVYGDNHADFIVESTRHRLPFVGRRTSGGRFSLHNGLMVLSKYPIVH 70
+++ NH DFI+ ST + G + + G++ H + Y +VH
Sbjct 468 DIFAMNHTDFIITSTFQEI--AGSKETVGQYESHTAFTLPGLYRVVH 512
> At3g02850
Length=828
Score = 29.3 bits (64), Expect = 8.7, Method: Composition-based stats.
Identities = 21/58 (36%), Positives = 31/58 (53%), Gaps = 11/58 (18%)
Query 22 LQEVYGDNHADFIVESTRHR---------LPFVGRRTSGGRFSLHNGLMVLSKYPIVH 70
L++ D+ D IVES +R L F G SGG+F++ NG+ +S+ IVH
Sbjct 21 LEDYEVDDFRDGIVESRGNRFNPLTNFLGLDFAG--GSGGKFTVINGIRDISRGSIVH 76
Lambda K H
0.326 0.141 0.440
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5080928138
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40