bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0487_orf1
Length=107
Score E
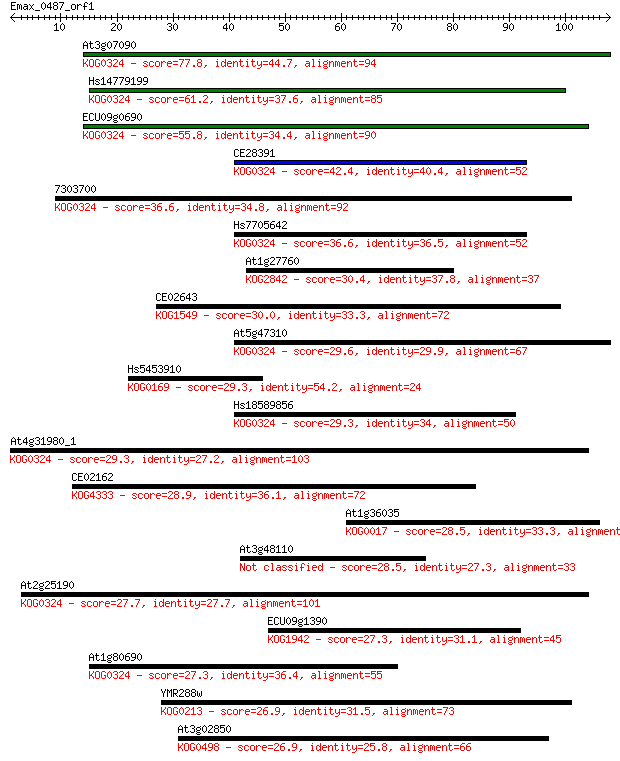
Sequences producing significant alignments: (Bits) Value
At3g07090 77.8 4e-15
Hs14779199 61.2 4e-10
ECU09g0690 55.8 2e-08
CE28391 42.4 2e-04
7303700 36.6 0.012
Hs7705642 36.6 0.012
At1g27760 30.4 0.72
CE02643 30.0 1.0
At5g47310 29.6 1.5
Hs5453910 29.3 1.6
Hs18589856 29.3 1.8
At4g31980_1 29.3 2.1
CE02162 28.9 2.7
At1g36035 28.5 2.9
At3g48110 28.5 3.2
At2g25190 27.7 5.2
ECU09g1390 27.3 6.4
At1g80690 27.3 7.9
YMR288w 26.9 9.1
At3g02850 26.9 9.8
> At3g07090
Length=265
Score = 77.8 bits (190), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 42/94 (44%), Positives = 63/94 (67%), Gaps = 3/94 (3%)
Query 14 RVILRVYDISRGLASRLAPMLLGRPLEGVWHSGIQLGDLEYFYGGGVVRMRPYEIESMYG 73
+V L VYD+S+GLA +L+ LLG+ +EGVWH+GI + EYF+GGG+ + P + YG
Sbjct 7 KVTLNVYDLSQGLARQLSQSLLGKVIEGVWHTGIVVYGNEYFFGGGIQHL-PVG-RTPYG 64
Query 74 MSPNKIEELGVTHKTNQEIQTFLTSISPRFTAET 107
+P + ELG++H + +L ISPR+TAE+
Sbjct 65 -TPIRTIELGLSHVPKDVFEMYLEEISPRYTAES 97
> Hs14779199
Length=168
Score = 61.2 bits (147), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 32/85 (37%), Positives = 54/85 (63%), Gaps = 2/85 (2%)
Query 15 VILRVYDISRGLASRLAPMLLGRPLEGVWHSGIQLGDLEYFYGGGVVRMRPYEIESMYGM 74
V L VYD+S+GLA RL+P++LG+ LEG+WH+ I + E+F+G G + P ++ G
Sbjct 9 VKLYVYDLSKGLARRLSPIMLGKQLEGIWHTSIVVHKDEFFFGSGGISSCP-PGGTLLG- 66
Query 75 SPNKIEELGVTHKTNQEIQTFLTSI 99
P+ + ++G T T + +L+S+
Sbjct 67 PPDSVVDVGSTEVTEEIFLEYLSSL 91
> ECU09g0690
Length=150
Score = 55.8 bits (133), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 31/90 (34%), Positives = 48/90 (53%), Gaps = 3/90 (3%)
Query 14 RVILRVYDISRGLASRLAPMLLGRPLEGVWHSGIQLGDLEYFYGGGVVRMRPYEIESMYG 73
VILRVY + + R LG+ +WH+ I++ EYF+ G+++ RP ++YG
Sbjct 5 NVILRVYSLGDEVLKRFITSSLGKSEACIWHTSIEVYGTEYFFQNGIMKARPG--STIYG 62
Query 74 MSPNKIEELGVTHKTNQEIQTFLTSISPRF 103
+P KI +LG T + FL SI+ F
Sbjct 63 -TPLKIHDLGATDIPEVVFEDFLFSIAEDF 91
> CE28391
Length=334
Score = 42.4 bits (98), Expect = 2e-04, Method: Composition-based stats.
Identities = 21/52 (40%), Positives = 30/52 (57%), Gaps = 5/52 (9%)
Query 41 GVWHSGIQLGDLEYFYGGGVVRMRPYEIESMYGMSPNKIEELGVTHKTNQEI 92
G++HSGI++ +EY YGG PY+ ++ SP EELG T K + I
Sbjct 52 GIFHSGIEVFGVEYAYGG-----HPYQFSGVFENSPQDAEELGETFKFKESI 98
> 7303700
Length=205
Score = 36.6 bits (83), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 32/102 (31%), Positives = 45/102 (44%), Gaps = 29/102 (28%)
Query 9 SSSGTR--VILRVYDI--------SRGLASRLAPMLLGRPLEGVWHSGIQLGDLEYFYGG 58
S+ GTR VIL VYD+ S GL GV+HSG++ E+ YGG
Sbjct 31 SNMGTREPVILNVYDMYWINEYTTSIGL--------------GVFHSGVEAFGTEFAYGG 76
Query 59 GVVRMRPYEIESMYGMSPNKIEELGVTHKTNQEIQTFLTSIS 100
P+ ++ +SP +ELG + Q IQ T +
Sbjct 77 -----HPFPFTGVFEISPRDHDELGDQFQFRQSIQIGCTDFT 113
> Hs7705642
Length=193
Score = 36.6 bits (83), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 19/52 (36%), Positives = 28/52 (53%), Gaps = 5/52 (9%)
Query 41 GVWHSGIQLGDLEYFYGGGVVRMRPYEIESMYGMSPNKIEELGVTHKTNQEI 92
GV+HSGI++ E+ YGG PY ++ +SP ELG T K + +
Sbjct 27 GVFHSGIEVYGREFAYGG-----HPYPFSGIFEISPGNASELGETFKFKEAV 73
> At1g27760
Length=459
Score = 30.4 bits (67), Expect = 0.72, Method: Compositional matrix adjust.
Identities = 14/39 (35%), Positives = 24/39 (61%), Gaps = 2/39 (5%)
Query 43 WHSGIQLGDLEYFYGGGVVR-MRPYE-IESMYGMSPNKI 79
W+ IQL L++F GGG ++ M+ E + ++ +P KI
Sbjct 370 WYQMIQLNYLKHFLGGGFIKHMQENEFLHDVFSFTPKKI 408
> CE02643
Length=328
Score = 30.0 bits (66), Expect = 1.0, Method: Composition-based stats.
Identities = 24/79 (30%), Positives = 34/79 (43%), Gaps = 12/79 (15%)
Query 27 ASRLAPMLLGRPLEGVWHSGIQ-------LGDLEYFYGGGVVRMRPYEIESMYGMSPNKI 79
+ R+ PMLLG E W SG + LG+ Y G + IES+ + +
Sbjct 169 SKRIPPMLLGGNQESGWRSGTENTPMIVGLGEAAKVYNEGFL-----NIESILRQNRDYF 223
Query 80 EELGVTHKTNQEIQTFLTS 98
EEL + N I F+ S
Sbjct 224 EELLLKRLRNSHIIHFIGS 242
> At5g47310
Length=245
Score = 29.6 bits (65), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 20/73 (27%), Positives = 34/73 (46%), Gaps = 9/73 (12%)
Query 41 GVWHSGIQLGDLEYFYGG------GVVRMRPYEIESMYGMSPNKIEELGVTHKTNQEIQT 94
G++HSGI+ EY YG GV + P S G + LG T + + ++
Sbjct 50 GIFHSGIEAHGFEYGYGAHEYSSSGVFEVEP---RSCPGFIFRRSVLLGTTSMSRSDFRS 106
Query 95 FLTSISPRFTAET 107
F+ +S ++ +T
Sbjct 107 FMEKLSRKYHGDT 119
> Hs5453910
Length=756
Score = 29.3 bits (64), Expect = 1.6, Method: Composition-based stats.
Identities = 13/24 (54%), Positives = 18/24 (75%), Gaps = 0/24 (0%)
Query 22 ISRGLASRLAPMLLGRPLEGVWHS 45
++R L + L PMLL RPL+GV +S
Sbjct 401 MARHLHAILGPMLLNRPLDGVTNS 424
> Hs18589856
Length=154
Score = 29.3 bits (64), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 17/50 (34%), Positives = 25/50 (50%), Gaps = 5/50 (10%)
Query 41 GVWHSGIQLGDLEYFYGGGVVRMRPYEIESMYGMSPNKIEELGVTHKTNQ 90
GV+HS I++ E+ YGG PY ++ +SP ELG K +
Sbjct 27 GVFHSEIEVYGREFAYGG-----HPYPFSGIFEISPGNASELGEPFKFKE 71
> At4g31980_1
Length=266
Score = 29.3 bits (64), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 28/111 (25%), Positives = 51/111 (45%), Gaps = 19/111 (17%)
Query 1 HAAAKMANSSSGTRVILRVYDISRGLASRLAPM-LLGRPLE-GVWHSGIQLGDLEYFYGG 58
+ + + +S V L VYD L PM + G L G++HSG+++ +EY YG
Sbjct 5 NVSVRRKKNSGTVPVYLNVYD--------LTPMNVYGYWLGIGIYHSGLEVHGVEYGYGA 56
Query 59 ------GVVRMRPYEIESMYGMSPNKIEELGVTHKTNQEIQTFLTSISPRF 103
G+ + P + G + K +G T +E+++F+ +S +
Sbjct 57 HEKSSSGIFEVEPKKCP---GFTFRKSILVGETEMKAKEVRSFMEKLSEEY 104
> CE02162
Length=380
Score = 28.9 bits (63), Expect = 2.7, Method: Composition-based stats.
Identities = 26/87 (29%), Positives = 36/87 (41%), Gaps = 17/87 (19%)
Query 12 GTRVILRVYDISRGLASRLAPMLLGRPLEGVWHSGIQLG---------------DLEYFY 56
G RV +YD+S AS L+ L P E +W + L L+ F
Sbjct 38 GRRVQENMYDVSWDQASELSYTYLDTPSEHIWTPIVPLNATTVPVTCGYVKGIMHLKLFR 97
Query 57 GGGVVRMRPYEIESMYGMSPNKIEELG 83
G +R E ES Y ++P + ELG
Sbjct 98 CPG-IRQACIEYESQY-LTPKQFTELG 122
> At1g36035
Length=1418
Score = 28.5 bits (62), Expect = 2.9, Method: Composition-based stats.
Identities = 15/49 (30%), Positives = 24/49 (48%), Gaps = 4/49 (8%)
Query 61 VRMRPYE----IESMYGMSPNKIEELGVTHKTNQEIQTFLTSISPRFTA 105
++M PYE S N+ E LG T+K + ++ L + P+F A
Sbjct 135 LKMEPYETIVKFSSKISALANEAEVLGKTYKDQKLVKKLLRCLPPKFPA 183
> At3g48110
Length=1067
Score = 28.5 bits (62), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 9/33 (27%), Positives = 17/33 (51%), Gaps = 0/33 (0%)
Query 42 VWHSGIQLGDLEYFYGGGVVRMRPYEIESMYGM 74
+W G+++ YF G + + P +E YG+
Sbjct 199 IWMDGMEITQFTYFQQAGSLPLSPVSVEITYGL 231
> At2g25190
Length=240
Score = 27.7 bits (60), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 28/112 (25%), Positives = 47/112 (41%), Gaps = 25/112 (22%)
Query 3 AAKMANSSSGTRVILRVYDISRGLASRLAPM-----LLGRPLEGVWHSGIQLGDLEYFYG 57
+ K S V L VYD L PM LG GV+HSG+++ +EY +G
Sbjct 7 SVKRKKQSGSVPVYLNVYD--------LTPMNAYGYWLGL---GVFHSGVEVHGVEYAFG 55
Query 58 G------GVVRMRPYEIESMYGMSPNKIEELGVTHKTNQEIQTFLTSISPRF 103
G+ + P + G + K +G T +E++ F+ ++ +
Sbjct 56 AHESSSTGIFEVEPKKCP---GFTFRKSILVGKTDLVAKEVRVFMEKLAEEY 104
> ECU09g1390
Length=426
Score = 27.3 bits (59), Expect = 6.4, Method: Compositional matrix adjust.
Identities = 14/45 (31%), Positives = 27/45 (60%), Gaps = 0/45 (0%)
Query 47 IQLGDLEYFYGGGVVRMRPYEIESMYGMSPNKIEELGVTHKTNQE 91
+++ +L+ Y G VV +R + E+ P +I+E+ V KT++E
Sbjct 114 VRMRELKDVYEGEVVELRIVDEENPLSSYPKRIKEMFVILKTSKE 158
> At1g80690
Length=231
Score = 27.3 bits (59), Expect = 7.9, Method: Compositional matrix adjust.
Identities = 20/63 (31%), Positives = 29/63 (46%), Gaps = 16/63 (25%)
Query 15 VILRVYDIS--RGLASRLAPMLLGRPLEGVWHSGIQLGDLEYFYGG------GVVRMRPY 66
V L VYD++ G A L GV+HSG+++ +EY YG G+ P
Sbjct 17 VYLNVYDLTPINGYAYWLG--------LGVYHSGVEVHGIEYAYGAHEYPSTGIFEGEPK 68
Query 67 EIE 69
+ E
Sbjct 69 QCE 71
> YMR288w
Length=971
Score = 26.9 bits (58), Expect = 9.1, Method: Composition-based stats.
Identities = 23/74 (31%), Positives = 38/74 (51%), Gaps = 3/74 (4%)
Query 28 SRLAPMLLGRPLEGV-WHSGIQLGDLEYFYGGGVVRMRPYEIESMYGMSPNKIEELGVTH 86
+RL P+LL R LE H I+ D + G + + PY + + +P I+E +
Sbjct 202 NRLLPILLDRSLEDQERHLMIKTIDRVLYQLGDLTK--PYVHKILVVAAPLLIDEDPMVR 259
Query 87 KTNQEIQTFLTSIS 100
T QEI T L++++
Sbjct 260 STGQEIITNLSTVA 273
> At3g02850
Length=828
Score = 26.9 bits (58), Expect = 9.8, Method: Compositional matrix adjust.
Identities = 17/66 (25%), Positives = 33/66 (50%), Gaps = 1/66 (1%)
Query 31 APMLLGRPLEGVWHSGIQLGDLEYFYGGGVVRMRPYEIESMYGMSPNKIEELGVTHKTNQ 90
P+ G E + I+L + E+F G V+ + ++ +Y + +EE+G+T ++
Sbjct 401 VPLFRGCSSEFINQIVIRLHE-EFFLPGEVIMEQGSVVDQLYFVCHGVLEEIGITKDGSE 459
Query 91 EIQTFL 96
EI L
Sbjct 460 EIVAVL 465
Lambda K H
0.318 0.134 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1164169380
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40