bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0508_orf1
Length=181
Score E
Sequences producing significant alignments: (Bits) Value
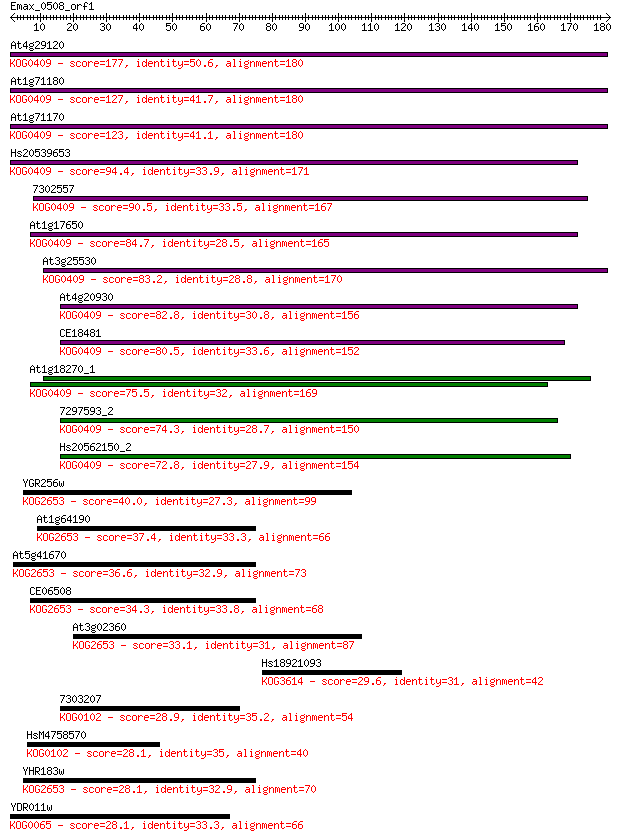
At4g29120 177 1e-44
At1g71180 127 9e-30
At1g71170 123 2e-28
Hs20539653 94.4 1e-19
7302557 90.5 2e-18
At1g17650 84.7 1e-16
At3g25530 83.2 2e-16
At4g20930 82.8 3e-16
CE18481 80.5 1e-15
At1g18270_1 75.5 5e-14
7297593_2 74.3 1e-13
Hs20562150_2 72.8 3e-13
YGR256w 40.0 0.002
At1g64190 37.4 0.015
At5g41670 36.6 0.030
CE06508 34.3 0.12
At3g02360 33.1 0.33
Hs18921093 29.6 3.9
7303207 28.9 5.3
HsM4758570 28.1 8.8
YHR183w 28.1 8.9
YDR011w 28.1 9.1
> At4g29120
Length=334
Score = 177 bits (448), Expect = 1e-44, Method: Compositional matrix adjust.
Identities = 91/180 (50%), Positives = 119/180 (66%), Gaps = 1/180 (0%)
Query 1 AEEVYVHLKEKGVAAVDAPVTGGDVGARNGTLTVMAGGDAKTFENLRPLFEAVGTNIIHA 60
AEE+ K ++DAPV+GGD+GA+NG L++ AGGD T + L PLF +G +
Sbjct 139 AEEIAKAASFKNCFSIDAPVSGGDLGAKNGKLSIFAGGDETTVKRLDPLFSLMG-KVNFM 197
Query 61 GPAGSGQRMKLCNQITLCTNLVGLAEGLLFAERAGLDIEKTLAVLSGGGAASWSVSTYGP 120
G +G GQ KL NQIT+ + ++GL EGL++A +AGLD++K L +S G A S S+ YG
Sbjct 198 GTSGKGQFAKLANQITIASTMLGLVEGLIYAHKAGLDVKKFLEAISTGAAGSKSIDLYGD 257
Query 121 RIRDGDFRPGFFVRHLVKDLRLALNEAARLKLALPGLGLALQLYQALEAQGGGSLGVHAL 180
RI DF PGF+V H VKDL + LNE R+ LALPGL LA QLY +L+A G G LG AL
Sbjct 258 RILKRDFDPGFYVNHFVKDLGICLNECQRMGLALPGLALAQQLYLSLKAHGEGDLGTQAL 317
> At1g71180
Length=297
Score = 127 bits (320), Expect = 9e-30, Method: Compositional matrix adjust.
Identities = 75/180 (41%), Positives = 105/180 (58%), Gaps = 7/180 (3%)
Query 1 AEEVYVHLKEKGVAAVDAPVTGGDVGARNGTLTVMAGGDAKTFENLRPLFEAVGTNIIHA 60
A E++ + + AVDAPV+GGD GAR GTL + AGGD++ E L P+ + +GT + +
Sbjct 114 AREIHAEARRRNCWAVDAPVSGGDAGAREGTLGIFAGGDSEIVEWLSPVMKNIGT-VTYM 172
Query 61 GPAGSGQRMKLCNQITLCTNLVGLAEGLLFAERAGLDIEKTLAVLSGGGAASWSVSTYGP 120
G AGSGQ K+ NQI +NLVGLAEG++FAE+AGLD K L + G A S + +G
Sbjct 173 GEAGSGQSCKIGNQIAGASNLVGLAEGIVFAEKAGLDTVKWLEAVKDGAAGSAVMRLFGE 232
Query 121 RIRDGDFRPGFFVRHLVKDLRLALNEAARLKLALPGLGLALQLYQALEAQGGGSLGVHAL 180
I D+R F ++VKDL +A A L+ QL+ + A G G LG+ +
Sbjct 233 MIVKRDYRATGFAEYMVKDLGMAAEAAMPGAA------LSKQLFTGMVANGDGKLGIQGV 286
> At1g71170
Length=299
Score = 123 bits (309), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 74/180 (41%), Positives = 106/180 (58%), Gaps = 5/180 (2%)
Query 1 AEEVYVHLKEKGVAAVDAPVTGGDVGARNGTLTVMAGGDAKTFENLRPLFEAVGTNIIHA 60
A E+Y + + AVDAPV+GGD GAR G LT+ AGGD++ E L P+ + +G +
Sbjct 114 AREIYAEARRRDCWAVDAPVSGGDAGAREGKLTIFAGGDSEIVEWLAPVMKTMGI-VRFM 172
Query 61 GPAGSGQRMKLCNQITLCTNLVGLAEGLLFAERAGLDIEKTLAVLSGGGAASWSVSTYGP 120
G AGSGQ K+ NQI + +N++GLAEG++FAE+AGLD K L + G A S + +G
Sbjct 173 GGAGSGQSCKIGNQICVGSNMIGLAEGIVFAEKAGLDPVKWLEAVKDGAAGSAVMRLFGE 232
Query 121 RIRDGDFRPGFFVRHLVKDLRLALNEAARLKLALPGLGLALQLYQALEAQGGGSLGVHAL 180
+ D++ F ++VKD L AA +A+PG L QL+ + A G G LG +
Sbjct 233 MMAVRDYKATGFAEYMVKD----LGMAAEAAMAMPGTALNKQLFTVMVANGDGKLGFQGV 288
> Hs20539653
Length=336
Score = 94.4 bits (233), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 58/178 (32%), Positives = 94/178 (52%), Gaps = 7/178 (3%)
Query 1 AEEVYVHLKEKGVAAVDAPVTGGDVGARNGTLTVMAGGDAKTFENLRPLFEAVGTNIIHA 60
++E+ +++ G +DAPV+GG AR+G LT M GG F + L +G+N+++
Sbjct 140 SKELAKEVEKMGAVFMDAPVSGGVGAARSGNLTFMVGGVEDEFAAAQELLGCMGSNVVYC 199
Query 61 GPAGSGQRMKLCNQITLCTNLVGLAEGLLFAERAGLDIEKTLAVLSGGGAASWSVSTYGP 120
G G+GQ K+CN + L +++G AE + R GLD + +L+ WS TY P
Sbjct 200 GAVGTGQAAKICNNMLLAISMIGTAEAMNLGIRLGLDPKLLAKILNMSSGRCWSSDTYNP 259
Query 121 R--IRDG-----DFRPGFFVRHLVKDLRLALNEAARLKLALPGLGLALQLYQALEAQG 171
+ DG +++ GF + KDL LA + A K + LA Q+Y+ + A+G
Sbjct 260 VPGVMDGVPSANNYQGGFGTTLMAKDLGLAQDSATSTKSPILLGSLAHQIYRMMCAKG 317
> 7302557
Length=315
Score = 90.5 bits (223), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 56/174 (32%), Positives = 84/174 (48%), Gaps = 7/174 (4%)
Query 8 LKEKGVAAVDAPVTGGDVGARNGTLTVMAGGDAKTFENLRPLFEAVGTNIIHAGPAGSGQ 67
+ KG +DAPV+GG GA TLT M GG + ++ + E +G I H G G GQ
Sbjct 125 ISAKGARFIDAPVSGGVPGAEQATLTFMVGGTEAEYNAVKAVLECMGKKITHCGVYGMGQ 184
Query 68 RMKLCNQITLCTNLVGLAEGLLFAERAGLDIEKTLAVLSGGGAASWSVSTYGP------- 120
KLCN + L +++G++E + A R GLD +++ W+ Y P
Sbjct 185 AAKLCNNMMLAISMIGVSEAMNLAVRQGLDANVFAEIINSSTGRCWASEIYNPVPGVCPS 244
Query 121 RIRDGDFRPGFFVRHLVKDLRLALNEAARLKLALPGLGLALQLYQALEAQGGGS 174
+ D+ GF + KDL LA A +P LA ++YQ+L +G G+
Sbjct 245 APANRDYAGGFSSALITKDLGLASGVANASNSPIPLGSLAHKVYQSLCDKGLGN 298
> At1g17650
Length=670
Score = 84.7 bits (208), Expect = 1e-16, Method: Composition-based stats.
Identities = 47/165 (28%), Positives = 82/165 (49%), Gaps = 0/165 (0%)
Query 7 HLKEKGVAAVDAPVTGGDVGARNGTLTVMAGGDAKTFENLRPLFEAVGTNIIHAGPAGSG 66
+K+ G ++APV+G A +G L + GD +E P + +G + + G G+G
Sbjct 485 QIKDTGALFLEAPVSGSKKPAEDGQLIFLTAGDKPLYEKAAPFLDIMGKSKFYLGEVGNG 544
Query 67 QRMKLCNQITLCTNLVGLAEGLLFAERAGLDIEKTLAVLSGGGAASWSVSTYGPRIRDGD 126
MKL + + + + AEG+L +++ GLD + V+S G + S GP +
Sbjct 545 AAMKLVVNMIMGSMMASFAEGILLSQKVGLDPNVLVEVVSQGAINAPMYSLKGPSMIKSV 604
Query 127 FRPGFFVRHLVKDLRLALNEAARLKLALPGLGLALQLYQALEAQG 171
+ F ++H KD+RLAL A + + P A +LY+ ++ G
Sbjct 605 YPTAFPLKHQQKDMRLALGLAESVSQSTPIAAAANELYKVAKSYG 649
> At3g25530
Length=289
Score = 83.2 bits (204), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 49/170 (28%), Positives = 84/170 (49%), Gaps = 0/170 (0%)
Query 11 KGVAAVDAPVTGGDVGARNGTLTVMAGGDAKTFENLRPLFEAVGTNIIHAGPAGSGQRMK 70
KG V+ PV+G A +G L ++A GD FE P F+ +G + G G+G +MK
Sbjct 111 KGGRFVEGPVSGSKKPAEDGQLIILAAGDKALFEESIPAFDVLGKRSFYLGQVGNGAKMK 170
Query 71 LCNQITLCTNLVGLAEGLLFAERAGLDIEKTLAVLSGGGAASWSVSTYGPRIRDGDFRPG 130
L + + + + +EGL+ A+++GL + L +L G + GP + + P
Sbjct 171 LIVNMIMGSMMNAFSEGLVLADKSGLSSDTLLDILDLGAMTNPMFKGKGPSMNKSSYPPA 230
Query 131 FFVRHLVKDLRLALNEAARLKLALPGLGLALQLYQALEAQGGGSLGVHAL 180
F ++H KD+RLAL +++P A + ++ + G G L A+
Sbjct 231 FPLKHQQKDMRLALALGDENAVSMPVAAAANEAFKKARSLGLGDLDFSAV 280
> At4g20930
Length=371
Score = 82.8 bits (203), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 48/163 (29%), Positives = 80/163 (49%), Gaps = 7/163 (4%)
Query 16 VDAPVTGGDVGARNGTLTVMAGGDAKTFENLRPLFEAVGTNIIHAGPAGSGQRMKLCNQI 75
+DAPV+GG + A GTLT M GG + RP+ +++G I+ G +G+G K+CN +
Sbjct 189 LDAPVSGGVLAAEAGTLTFMVGGPEDAYLAARPILQSMGRTSIYCGGSGNGSAAKICNNL 248
Query 76 TLCTNLVGLAEGLLFAERAGLDIEKTLAVLSGGGAASWSVSTYGPR--IRDG-----DFR 128
+ +++G +E L + G+ VL+ WS Y P + G D+
Sbjct 249 AMAVSMLGTSEALALGQSLGISASTLTEVLNTSSGRCWSSDAYNPVPGVMKGVPSSRDYN 308
Query 129 PGFFVRHLVKDLRLALNEAARLKLALPGLGLALQLYQALEAQG 171
GF + + KDL LA A + P + A ++Y+ + +G
Sbjct 309 GGFASKLMAKDLNLAAASAEEVGHKSPLISKAQEIYKKMCEEG 351
> CE18481
Length=299
Score = 80.5 bits (197), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 51/160 (31%), Positives = 77/160 (48%), Gaps = 8/160 (5%)
Query 16 VDAPVTGGDVGARNGTLTVMAG-GDAKTFENLRPLFEAVGTNIIHAGPAGSGQRMKLCNQ 74
+DAP++GG GA+ TLT M G G+ TF+ + +G NI++ G G+G K+CN
Sbjct 117 IDAPISGGVTGAQQATLTFMVGAGNDATFKRAEAVLSLMGKNIVNLGAVGNGTAAKICNN 176
Query 75 ITLCTNLVGLAEGLLFAERAGLDIEKTLAVLSGGGAASWSVSTYGPRIRDGDFRP----- 129
+ L +V +AE + GLD + +++ WS TY P + P
Sbjct 177 MLLGIQMVAVAETMNLGISMGLDAKALAGIVNTSSGRCWSSDTYNPVPGVIENIPSCRGY 236
Query 130 --GFFVRHLVKDLRLALNEAARLKLALPGLGLALQLYQAL 167
GF + KDL LA N + + P LA Q+Y+ L
Sbjct 237 AGGFGTTLMAKDLSLAQNASTNTQAPTPMGSLAHQIYRIL 276
> At1g18270_1
Length=1202
Score = 75.5 bits (184), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 52/166 (31%), Positives = 80/166 (48%), Gaps = 1/166 (0%)
Query 11 KGVAAVDAPVTGGDVGARNGTLTVMAGGDAKTFENLRPLFEAVGTNI-IHAGPAGSGQRM 69
K + VDAPV+GG A G LT+MA G + ++ + A+ + + G G+G +
Sbjct 442 KDLKLVDAPVSGGVKRAAMGELTIMASGTDEALKSAGLVLSALSEKLYVIKGGCGAGSGV 501
Query 70 KLCNQITLCTNLVGLAEGLLFAERAGLDIEKTLAVLSGGGAASWSVSTYGPRIRDGDFRP 129
K+ NQ+ ++ AE + F R GL+ K V+S G SW P + D D+ P
Sbjct 502 KMVNQLLAGVHIASAAEAMAFGARLGLNTRKLFNVISNSGGTSWMFENRVPHMLDNDYTP 561
Query 130 GFFVRHLVKDLRLALNEAARLKLALPGLGLALQLYQALEAQGGGSL 175
+ VKDL + E + K+ L +A QL+ A A G G +
Sbjct 562 YSALDIFVKDLGIVTREGSSRKVPLHISTVAHQLFLAGSAAGWGRI 607
Score = 53.9 bits (128), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 39/157 (24%), Positives = 71/157 (45%), Gaps = 2/157 (1%)
Query 7 HLKEKGVAAVDAPVTGGDVGARNGTLTVMAGGDAKTFENLRPLFEAVGTNI-IHAGPAGS 65
H K + + VDA V G +G L ++A G + + +P A+ N+ G G+
Sbjct 119 HEKREQIFVVDAYVLKGMSELLDGKLMIIASGRSDSITRAQPYLTAMCQNLYTFEGEIGA 178
Query 66 GQRMKLCNQITLCTNLVGLAEGLLFAERAGLDIEKTLAVLSGGGAASWSVSTYGPRIRDG 125
G ++K+ N++ +LV E + +AG+ ++S SW + P +
Sbjct 179 GSKVKMVNELLEGIHLVAAVEAISLGSQAGVHPWILYDIISNAAGNSWIYKNHIPLLLKD 238
Query 126 DFRPGFFVRHLVKDLRLALNEAARLKLALPGLGLALQ 162
D G F+ L ++L + ++A L +P L +A Q
Sbjct 239 DIE-GRFLDVLSQNLAIVEDKAKSLPFPVPLLAVARQ 274
> 7297593_2
Length=494
Score = 74.3 bits (181), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 43/150 (28%), Positives = 71/150 (47%), Gaps = 0/150 (0%)
Query 16 VDAPVTGGDVGARNGTLTVMAGGDAKTFENLRPLFEAVGTNIIHAGPAGSGQRMKLCNQI 75
++A + G A G L ++AGGD FE F+ + N G G+ ++ L Q
Sbjct 324 LEAQIHGSRQEAAEGMLIILAGGDRSVFEECHSCFKTIAKNTFFLGNIGNACKVNLILQT 383
Query 76 TLCTNLVGLAEGLLFAERAGLDIEKTLAVLSGGGAASWSVSTYGPRIRDGDFRPGFFVRH 135
L +LVGLAE L A+R + + + + S + G + GDF P + H
Sbjct 384 ILGVSLVGLAEALALADRFSISLNDIIDIFDLTSMKSPMLLAKGKEMAKGDFNPQQPLSH 443
Query 136 LVKDLRLALNEAARLKLALPGLGLALQLYQ 165
+ +DLRL LN A L ++P + ++++
Sbjct 444 MQRDLRLVLNMAENLDQSMPVTSITNEVFK 473
> Hs20562150_2
Length=340
Score = 72.8 bits (177), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 43/154 (27%), Positives = 77/154 (50%), Gaps = 0/154 (0%)
Query 16 VDAPVTGGDVGARNGTLTVMAGGDAKTFENLRPLFEAVGTNIIHAGPAGSGQRMKLCNQI 75
++APV+G + +G L ++A GD +E+ F+A+G G G+ +M L +
Sbjct 170 LEAPVSGNQQLSNDGMLVILAAGDRGLYEDCSSCFQAMGKTSFFLGEVGNAAKMMLIVNM 229
Query 76 TLCTNLVGLAEGLLFAERAGLDIEKTLAVLSGGGAASWSVSTYGPRIRDGDFRPGFFVRH 135
+ + +AEGL A G + L +L+ G AS + I G+F+P F++++
Sbjct 230 VQGSFMATIAEGLTLAHVTGQSQQTLLDILNQGQLASIFLDQKCQNILQGNFKPDFYLKY 289
Query 136 LVKDLRLALNEAARLKLALPGLGLALQLYQALEA 169
+ KDLRLA+ + P A ++Y+ +A
Sbjct 290 IQKDLRLAIALGDAVNHPTPMAAAANEVYKRAKA 323
> YGR256w
Length=492
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 27/105 (25%), Positives = 54/105 (51%), Gaps = 7/105 (6%)
Query 5 YVHLKEKGVAAVDAPVTGGDVGARNGTLTVMAGGDAKTFENLRPLFEAVGTN------II 58
Y L ++G+ V + V+GG+ GAR G ++M GG A+ + +++ +F+++
Sbjct 115 YEELTKQGILFVGSGVSGGEDGARFGP-SLMPGGSAEAWPHIKNIFQSIAAKSNGEPCCE 173
Query 59 HAGPAGSGQRMKLCNQITLCTNLVGLAEGLLFAERAGLDIEKTLA 103
GPAGSG +K+ + ++ + E +R G +K ++
Sbjct 174 WVGPAGSGHYVKMVHNGIEYGDMQLICEAYDIMKRIGRFTDKEIS 218
> At1g64190
Length=487
Score = 37.4 bits (85), Expect = 0.015, Method: Compositional matrix adjust.
Identities = 22/72 (30%), Positives = 41/72 (56%), Gaps = 7/72 (9%)
Query 9 KEKGVAAVDAPVTGGDVGARNGTLTVMAGGDAKTFENLRPLFEAV------GTNIIHAGP 62
++KG+ + V+GG+ GARNG ++M GG + ++N++ + E V G + + G
Sbjct 122 EQKGLLYLGMGVSGGEEGARNGP-SLMPGGSFQAYDNIKDILEKVAAQVEDGPCVTYIGE 180
Query 63 AGSGQRMKLCNQ 74
GSG +K+ +
Sbjct 181 GGSGNFVKMVHN 192
> At5g41670
Length=487
Score = 36.6 bits (83), Expect = 0.030, Method: Compositional matrix adjust.
Identities = 24/79 (30%), Positives = 41/79 (51%), Gaps = 7/79 (8%)
Query 2 EEVYVHLKEKGVAAVDAPVTGGDVGARNGTLTVMAGGDAKTFENLRPLFEAV------GT 55
E V ++KG+ + V+GG+ GARNG ++M GG + N++ + E V G
Sbjct 115 ERRIVEAEKKGLLYLGMGVSGGEEGARNGP-SLMPGGSFTAYNNVKDILEKVAAQVEDGP 173
Query 56 NIIHAGPAGSGQRMKLCNQ 74
+ + G GSG +K+ +
Sbjct 174 CVTYIGEGGSGNFVKMVHN 192
> CE06508
Length=484
Score = 34.3 bits (77), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 23/74 (31%), Positives = 38/74 (51%), Gaps = 7/74 (9%)
Query 7 HLKEKGVAAVDAPVTGGDVGARNGTLTVMAGGDAKTFENLRPLFEAVGTN------IIHA 60
L KG+ V V+GG+ GAR G ++M GG+ K + +L+ +F+ +
Sbjct 115 QLAAKGIMFVGCGVSGGEEGARFGP-SLMPGGNPKAWPHLKDIFQKIAAKSNGEPCCDWV 173
Query 61 GPAGSGQRMKLCNQ 74
G AGSG +K+ +
Sbjct 174 GNAGSGHFVKMVHN 187
> At3g02360
Length=486
Score = 33.1 bits (74), Expect = 0.33, Method: Compositional matrix adjust.
Identities = 27/94 (28%), Positives = 45/94 (47%), Gaps = 16/94 (17%)
Query 20 VTGGDVGARNGTLTVMAGGDAKTFENLRPLFEAVGTNIIHAGPA-------GSGQRMKLC 72
V+GG+ GARNG ++M GG + ++N+ + V + +GP GSG
Sbjct 132 VSGGEEGARNGP-SMMPGGSYEAYKNIEDIVLKVAAQVRDSGPCVTYIGKGGSG------ 184
Query 73 NQITLCTNLVGLAEGLLFAERAGLDIEKTLAVLS 106
N + + N + + L AE D+ K++ LS
Sbjct 185 NFVKMVHNGIEYGDMQLIAE--AYDVLKSVGKLS 216
> Hs18921093
Length=2022
Score = 29.6 bits (65), Expect = 3.9, Method: Composition-based stats.
Identities = 13/42 (30%), Positives = 21/42 (50%), Gaps = 0/42 (0%)
Query 77 LCTNLVGLAEGLLFAERAGLDIEKTLAVLSGGGAASWSVSTY 118
+C NL+ G L + AG+D T++ G + WSV +
Sbjct 45 VCQNLIRCYCGRLIGDHAGIDYSWTISAAKGKESEQWSVEKH 86
> 7303207
Length=686
Score = 28.9 bits (63), Expect = 5.3, Method: Composition-based stats.
Identities = 19/54 (35%), Positives = 22/54 (40%), Gaps = 4/54 (7%)
Query 16 VDAPVTGGDVGARNGTLTVMAGGDAKTFENLRPLFEAVGTNIIHAGPAGSGQRM 69
V V G D+G N L VM G AK EN E T H G+R+
Sbjct 50 VKGAVIGIDLGTTNSCLAVMEGKQAKVIENA----EGARTTPSHVAFTKDGERL 99
> HsM4758570
Length=679
Score = 28.1 bits (61), Expect = 8.8, Method: Composition-based stats.
Identities = 14/40 (35%), Positives = 19/40 (47%), Gaps = 0/40 (0%)
Query 6 VHLKEKGVAAVDAPVTGGDVGARNGTLTVMAGGDAKTFEN 45
V ++ A+ V G D+G N + VM G AK EN
Sbjct 41 VSRRDYASEAIKGAVVGIDLGTTNSCVAVMEGKQAKVLEN 80
> YHR183w
Length=489
Score = 28.1 bits (61), Expect = 8.9, Method: Compositional matrix adjust.
Identities = 23/76 (30%), Positives = 43/76 (56%), Gaps = 7/76 (9%)
Query 5 YVHLKEKGVAAVDAPVTGGDVGARNGTLTVMAGGDAKTFENLRPLFEAVGTN------II 58
Y LK+KG+ V + V+GG+ GAR G ++M GG + + +++ +F+++
Sbjct 112 YEELKKKGILFVGSGVSGGEEGARYGP-SLMPGGSEEAWPHIKNIFQSISAKSDGEPCCE 170
Query 59 HAGPAGSGQRMKLCNQ 74
GPAG+G +K+ +
Sbjct 171 WVGPAGAGHYVKMVHN 186
> YDR011w
Length=1501
Score = 28.1 bits (61), Expect = 9.1, Method: Composition-based stats.
Identities = 22/81 (27%), Positives = 33/81 (40%), Gaps = 15/81 (18%)
Query 1 AEEVYVHLKEKGVAAVDAPVTGGDVGARNGT---------LTVMAGGDAKTFENLRPLFE 51
A+E +H+++ GV D G D A G LT+ G AK + +R +
Sbjct 118 ADEQGIHIRKAGVTIEDVSAKGVDASALEGATFGNILCLPLTIFKGIKAKRHQKMRQIIS 177
Query 52 AV------GTNIIHAGPAGSG 66
V G I+ G G+G
Sbjct 178 NVNALAEAGEMILVLGRPGAG 198
Lambda K H
0.319 0.138 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2842629034
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40