bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0530_orf2
Length=273
Score E
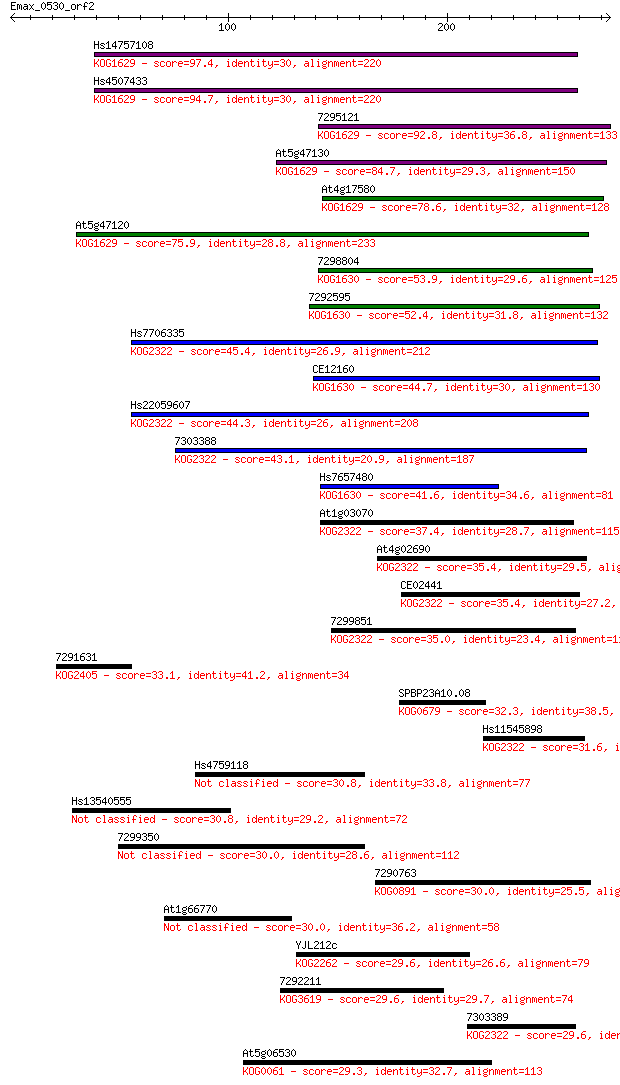
Sequences producing significant alignments: (Bits) Value
Hs14757108 97.4 3e-20
Hs4507433 94.7 2e-19
7295121 92.8 6e-19
At5g47130 84.7 2e-16
At4g17580 78.6 1e-14
At5g47120 75.9 8e-14
7298804 53.9 3e-07
7292595 52.4 1e-06
Hs7706335 45.4 1e-04
CE12160 44.7 2e-04
Hs22059607 44.3 3e-04
7303388 43.1 7e-04
Hs7657480 41.6 0.002
At1g03070 37.4 0.030
At4g02690 35.4 0.13
CE02441 35.4 0.14
7299851 35.0 0.15
7291631 33.1 0.64
SPBP23A10.08 32.3 1.1
Hs11545898 31.6 1.9
Hs4759118 30.8 2.9
Hs13540555 30.8 3.2
7299350 30.0 5.6
7290763 30.0 5.8
At1g66770 30.0 6.1
YJL212c 29.6 6.3
7292211 29.6 6.7
7303389 29.6 7.5
At5g06530 29.3 8.1
> Hs14757108
Length=237
Score = 97.4 bits (241), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 66/222 (29%), Positives = 127/222 (57%), Gaps = 9/222 (4%)
Query 39 SLLDFTPLSAPQKRHVSRIYAALTVNVLLTAVGVYGQLKWISLPPFL--SLMLSIGCVMG 96
+LL F+ ++ ++H+ ++YA+ + + + A G Y + + F+ L+ ++G ++
Sbjct 12 ALLKFSHITPSTQQHLKKVYASFALCMFVAAAGAYVHM----VTHFIQAGLLSALGSLIL 67
Query 97 LTYSSQKAHAESQMLTKERAVYFGGFGVLNGMLAANYLHAVHFYVGPQVIPAAFFASVAI 156
+ + H+ ++R GF L G+ L V P ++P AF + I
Sbjct 68 MIWLMATPHSHET--EQKRLGLLAGFAFLTGVGLGPALEFC-IAVNPSILPTAFMGTAMI 124
Query 157 FFCFSAAALVAKQRSYLYLGSILGAALTYLSLASLVNIFLRAQLVNNVILWGGLFMYLGF 216
F CF+ +AL A++RSYL+LG IL +AL+ L L+SL N+F + + L+ GL + GF
Sbjct 125 FTCFTLSALYARRRSYLFLGGILMSALSLLLLSSLGNVFFGSIWLFQANLYVGLVVMCGF 184
Query 217 VVYDTQLAVAQFDMGNRDYLLHALQFYVNFLSLFLRLVAILS 258
V++DTQL + + + G++DY+ H + +++F+++F +L+ IL+
Sbjct 185 VLFDTQLIIEKAEHGDQDYIWHCIDLFLDFITVFRKLMMILA 226
> Hs4507433
Length=237
Score = 94.7 bits (234), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 66/222 (29%), Positives = 125/222 (56%), Gaps = 9/222 (4%)
Query 39 SLLDFTPLSAPQKRHVSRIYAALTVNVLLTAVGVYGQLKWISLPPFL--SLMLSIGCVMG 96
+LL F+ ++ ++H+ ++YA+ + + + A G Y + + F+ L+ ++G ++
Sbjct 12 ALLKFSHITPSTQQHLKKVYASFALCMFVAAAGAYVHM----VTHFIQAGLLSALGSLIL 67
Query 97 LTYSSQKAHAESQMLTKERAVYFGGFGVLNGMLAANYLHAVHFYVGPQVIPAAFFASVAI 156
+ + H+ ++R GF L G+ L V P ++P AF + I
Sbjct 68 MIWLMATPHSHET--EQKRLGLLAGFAFLTGVGLGPALEFC-IAVNPSILPTAFMGTAMI 124
Query 157 FFCFSAAALVAKQRSYLYLGSILGAALTYLSLASLVNIFLRAQLVNNVILWGGLFMYLGF 216
F CF+ +AL A++RSYL+LG IL +AL+ L L+SL N+F + L+ GL + GF
Sbjct 125 FTCFTLSALYARRRSYLFLGGILMSALSLLLLSSLGNVFFGSIWPFQANLYVGLVVMCGF 184
Query 217 VVYDTQLAVAQFDMGNRDYLLHALQFYVNFLSLFLRLVAILS 258
V+ DTQL + + + G++DY+ H + +++F+++F +L+ IL+
Sbjct 185 VLVDTQLIIEKAEHGDQDYIWHCIDLFLDFITVFRKLMMILA 226
> 7295121
Length=245
Score = 92.8 bits (229), Expect = 6e-19, Method: Compositional matrix adjust.
Identities = 49/133 (36%), Positives = 81/133 (60%), Gaps = 3/133 (2%)
Query 141 VGPQVIPAAFFASVAIFFCFSAAALVAKQRSYLYLGSILGAALTYLSLASLVNIFLRAQL 200
+ P +I +A + F S +AL+A+Q YLYLG +L + + ++L SL N+ ++
Sbjct 112 INPAIILSALTGTFVTFISLSLSALLAEQGKYLYLGGMLVSVINTMALLSLFNMVFKSYF 171
Query 201 VNNVILWGGLFMYLGFVVYDTQLAVAQFDMGNRDYLLHALQFYVNFLSLFLRLVAILSER 260
V L+ G+F+ F+VYDTQ V + GNRD + HAL + + LS+F RL+ IL+++
Sbjct 172 VQVTQLYVGVFVMAAFIVYDTQNIVEKCRNGNRDVVQHALDLFFDVLSMFRRLLIILTQK 231
Query 261 QEENNRRKRERRE 273
+E R++ ERR+
Sbjct 232 EE---RKQNERRQ 241
> At5g47130
Length=187
Score = 84.7 bits (208), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 44/150 (29%), Positives = 84/150 (56%), Gaps = 3/150 (2%)
Query 122 FGVLNGMLAANYLHAVHFYVGPQVIPAAFFASVAIFFCFSAAALVAKQRSYLYLGSILGA 181
FGVL+G + + + ++ AF + IFFCFSA A++A++R Y+YLG +L +
Sbjct 40 FGVLHGASVGPCIKST-IDIDSSILITAFLGTAVIFFCFSAVAMLARRREYIYLGGLLSS 98
Query 182 ALTYLSLASLVNIFLRAQLVNNVILWGGLFMYLGFVVYDTQLAVAQFDMGNRDYLLHALQ 241
+ L+ + F A + + ++ GL +++G +V +TQ + + G+ DY +H+L
Sbjct 99 GFSLLTWLKNSDQFASATV--EIQMYLGLLLFVGCIVVNTQEIIEKAHCGDMDYAVHSLI 156
Query 242 FYVNFLSLFLRLVAILSERQEENNRRKRER 271
Y+ F+ +FL++++I+ + RR E
Sbjct 157 LYIGFVRVFLQILSIMWNTSADRIRRNNEE 186
> At4g17580
Length=247
Score = 78.6 bits (192), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 41/129 (31%), Positives = 73/129 (56%), Gaps = 1/129 (0%)
Query 143 PQVIPAAFFASVAIFFCFSAAALVAKQRSYLYLGSILGAALTYLSLASLVN-IFLRAQLV 201
V+ AF + F CFSAAA++A +R YLY G+ L ++ L + + IF + V
Sbjct 118 SSVLVTAFVGTAVAFVCFSAAAMLATRREYLYHGASLACCMSILWWVQIASSIFGGSTTV 177
Query 202 NNVILWGGLFMYLGFVVYDTQLAVAQFDMGNRDYLLHALQFYVNFLSLFLRLVAILSERQ 261
L+ GL +++G++V DTQ+ + G+ DY+ H+ F+ +F SLF++++ + R+
Sbjct 178 VKFELYFGLLIFVGYIVVDTQMITEKAHHGDMDYVQHSFTFFTDFASLFVQILVLNMFRK 237
Query 262 EENNRRKRE 270
+ R+ R
Sbjct 238 MKKGRKDRR 246
> At5g47120
Length=247
Score = 75.9 bits (185), Expect = 8e-14, Method: Compositional matrix adjust.
Identities = 67/236 (28%), Positives = 124/236 (52%), Gaps = 14/236 (5%)
Query 31 GASAVPARSLLDFTPLSAPQKRHVSRIYAALTVNVLLTAVGVYGQLKWISLPPFLSLMLS 90
G+ + SL +F +S + H+ R+Y L ++ +A G Y + W ++ +
Sbjct 13 GSRSWSYDSLKNFRQISPAVQNHLKRVYLTLCCALVASAFGAYLHVLW----NIGGILTT 68
Query 91 IGCVMGLTYSSQKAHAESQMLTKERAVYFGGFGVLNGMLAANYLHAVHFYVGPQVIPAAF 150
IGC+ + + E Q K ++ F VL G + L V V P ++ AF
Sbjct 69 IGCIGTMIWLLSCPPYEHQ---KRLSLLFVS-AVLEGA-SVGPLIKVAIDVDPSILITAF 123
Query 151 FASVAIFFCFSAAALVAKQRSYLYLGSILGAALT---YLSLASLVNIFLRAQLVNNVILW 207
+ F CFSAAA++A++R YLYLG +L + L+ +L AS +IF + + L+
Sbjct 124 VGTAIAFVCFSAAAMLARRREYLYLGGLLSSGLSMLMWLQFAS--SIFGGSASIFKFELY 181
Query 208 GGLFMYLGFVVYDTQLAVAQFDMGNRDYLLHALQFYVNFLSLFLRLVAILSERQEE 263
GL +++G++V DTQ + + +G+ DY+ H+L + +F+++F+R++ I+ + +
Sbjct 182 FGLLIFVGYMVVDTQEIIEKAHLGDMDYVKHSLTLFTDFVAVFVRILIIMLKNSAD 237
> 7298804
Length=305
Score = 53.9 bits (128), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 37/136 (27%), Positives = 67/136 (49%), Gaps = 11/136 (8%)
Query 141 VGPQVIPAAFFASVAIFFCFSAAALVAKQRSYLYLGSILGAALTYLSLASLVNIFLRAQL 200
+G ++ A + I S A A +L++G L L + +SL +++L
Sbjct 170 LGGPILTKALLYTSGIVGALSTVAACAPSEKFLHMGGPLAIGLGVVFASSLASMWLPPTT 229
Query 201 -----VNNVILWGGLFMYLGFVVYDTQLAV------AQFDMGNRDYLLHALQFYVNFLSL 249
+ ++ L+GGL ++ GF++YDTQ V Q+ D + HAL Y++ L++
Sbjct 230 AVGAGLASMSLYGGLILFSGFLLYDTQRIVKSAELYPQYSKFPYDPINHALAIYMDALNI 289
Query 250 FLRLVAILSERQEENN 265
F+R+ IL+ Q+ N
Sbjct 290 FIRIAIILAGDQKRKN 305
> 7292595
Length=341
Score = 52.4 bits (124), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 42/143 (29%), Positives = 70/143 (48%), Gaps = 16/143 (11%)
Query 137 VHFYVGPQVIPAAFFASVAIFFCFSAAALVAKQRSYLYLGSILGAALTYLSLASLVNIFL 196
+ F GP + AA + I S A A +LY+G L L + +SL +++L
Sbjct 204 ICFMGGPILTRAALYTG-GIVGGLSTIAACAPSDKFLYMGGPLAIGLGVVFASSLASMWL 262
Query 197 RAQL-----VNNVILWGGLFMYLGFVVYDTQLAV------AQFDMGNRDYLLHALQFYVN 245
+ ++ L+GGL ++ GF++YDTQ V Q+ D + ++ Y++
Sbjct 263 PPTTALGAGLASMSLYGGLVLFSGFLLYDTQRMVRRAEVYPQYSYTPYDPINASMSIYMD 322
Query 246 FLSLFLRLVAILSERQEENNRRK 268
L++F+R+V ILS Q RRK
Sbjct 323 VLNIFIRIVTILSGGQ----RRK 341
> Hs7706335
Length=258
Score = 45.4 bits (106), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 57/221 (25%), Positives = 105/221 (47%), Gaps = 28/221 (12%)
Query 56 RIYAALTVNVLLTAVGVYGQLKWISLPPFL----SLML-----SIGCVMGLTYSSQKAHA 106
++Y+ L++ VLLT V L + S+ F+ +L+L S+G + LT + K
Sbjct 57 KVYSILSLQVLLTTVTSTVFLYFESVRTFVHESPALILLFALGSLGLIFALTLNRHKYPL 116
Query 107 ESQMLTKERAVYFGGFGVLNGMLAANYLHAVHFYVGPQVIPAAFFASVAIFFCFSAAALV 166
+L GF +L + A V FY +I AF + +FF + L
Sbjct 117 NLYLLF--------GFTLLEALTVAV---VVTFY-DVYIILQAFILTTTVFFGLTVYTLQ 164
Query 167 AKQRSYLYLGSILGAALTYLSLASLVNIFLRAQLVNNVILWGGLFMYLGFVVYDTQLAVA 226
+K + + G+ L A L L L+ + F ++++ V+ G ++ GF++YDT +
Sbjct 165 SK-KDFSKFGAGLFALLWILCLSGFLKFFFYSEIMELVLAAAGALLFCGFIIYDTHSLMH 223
Query 227 QFDMGNRDYLLHALQFYVNFLSLFLRLVAILSERQEENNRR 267
+ + +Y+L A+ Y++ ++LFL L+ L E N++
Sbjct 224 K--LSPEEYVLAAISLYLDIINLFLHLLRFL----EAVNKK 258
> CE12160
Length=342
Score = 44.7 bits (104), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 39/149 (26%), Positives = 69/149 (46%), Gaps = 22/149 (14%)
Query 139 FYVGPQVIPAAFFASVAIFFCFSAAALVAKQRSYLYLGSILGAALTYLSLASLVNIFLR- 197
F GP + AA++ + I SA A+ A +L + L + +A++ FL
Sbjct 197 FMAGPVLTRAAWY-TAGIVGGLSATAITAPSEKFLMMSGPLAMGFGVVFVANIGAFFLPP 255
Query 198 ----AQLVNNVILWGGLFMYLGFVVYDTQLAVAQF------------DMGNR--DYLLHA 239
+ +++++GGL ++ F++YDTQ V + DM R D +
Sbjct 256 GSALGASLASIVVYGGLILFSAFLLYDTQRLVKKAENHPHSSQLYGSDMQIRSFDPINAQ 315
Query 240 LQFYVNFLSLFLRLVAILSERQEENNRRK 268
+ Y++ L++F+RLV I+ NRRK
Sbjct 316 MSIYMDVLNIFMRLVMIMGGM--NGNRRK 342
> Hs22059607
Length=285
Score = 44.3 bits (103), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 54/217 (24%), Positives = 102/217 (47%), Gaps = 24/217 (11%)
Query 56 RIYAALTVNVLLTAVGVYGQLKWISLPPFL----SLML-----SIGCVMGLTYSSQKAHA 106
++Y+ L++ VLLT V L + S+ F+ +L+L S+G + L + K
Sbjct 84 KVYSILSLQVLLTTVTSTVFLYFESVRTFVHESPALILLFALGSLGLIFALILNRHKYPL 143
Query 107 ESQMLTKERAVYFGGFGVLNGMLAANYLHAVHFYVGPQVIPAAFFASVAIFFCFSAAALV 166
+L GF +L + A V FY +I AF + +FF + L
Sbjct 144 NLYLLF--------GFTLLEALTVAV---VVTFY-DVYIILQAFILTTTVFFGLTVYTLQ 191
Query 167 AKQRSYLYLGSILGAALTYLSLASLVNIFLRAQLVNNVILWGGLFMYLGFVVYDTQLAVA 226
+K + + G+ L A L L L+ + F ++++ V+ G ++ GF++YDT +
Sbjct 192 SK-KDFSKFGAGLFALLWILCLSGFLKFFFYSEIMELVLAAAGALLFCGFIIYDTHSLMH 250
Query 227 QFDMGNRDYLLHALQFYVNFLSLFLRLVAILSERQEE 263
+ + +Y+L A+ Y++ ++LFL L+ L ++
Sbjct 251 K--LSPEEYVLAAISLYLDIINLFLHLLRFLEAVNKK 285
> 7303388
Length=324
Score = 43.1 bits (100), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 39/190 (20%), Positives = 85/190 (44%), Gaps = 19/190 (10%)
Query 76 LKWISLPPFLSLMLSIGCVMGLTYSSQKAHAESQMLTKERAVYFGGFGVLNGMLAANYLH 135
L W++L L MLS+ C + + + T ++ L G+ A Y
Sbjct 151 LFWVALGVMLVTMLSMACCESVRRQTPTNFIFLGLFTAAQSF-------LMGVSATKY-- 201
Query 136 AVHFYVGPQVIPAAFFASVAIFFCFSAAALVAKQRSYLYLGSILGAALTYLSLASLVNIF 195
P+ + A + A+ + AL K + +G IL A + + +V IF
Sbjct 202 ------APKEVLMAVGITAAVCLALTIFALQTK-YDFTMMGGILIACMVVFLIFGIVAIF 254
Query 196 LRAQLVNNVILWGGLFMYLGFVVYDTQLAVA---QFDMGNRDYLLHALQFYVNFLSLFLR 252
++ +++ V G ++ +++YDTQL + ++ + +Y+ AL Y++ +++F+
Sbjct 255 VKGKIITLVYASIGALLFSVYLIYDTQLMMGGEHKYSISPEEYIFAALNLYLDIINIFMY 314
Query 253 LVAILSERQE 262
++ I+ ++
Sbjct 315 ILTIIGASRD 324
> Hs7657480
Length=319
Score = 41.6 bits (96), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 28/86 (32%), Positives = 48/86 (55%), Gaps = 6/86 (6%)
Query 142 GPQVIPAAFFASVAIFFCFSAAALVAKQRSYLYLGSILGAALTYLSLASLVNIFLRAQLV 201
GP +I AA++ + I S A+ A +L +G+ LG L + ++SL ++FL V
Sbjct 212 GPLLIRAAWY-TAGIVGGLSTVAMCAPSEKFLNMGAPLGVGLGLVFVSSLGSMFLPPTTV 270
Query 202 N-----NVILWGGLFMYLGFVVYDTQ 222
+V ++GGL ++ F++YDTQ
Sbjct 271 AGATLYSVAMYGGLVLFSMFLLYDTQ 296
> At1g03070
Length=247
Score = 37.4 bits (85), Expect = 0.030, Method: Compositional matrix adjust.
Identities = 33/121 (27%), Positives = 63/121 (52%), Gaps = 12/121 (9%)
Query 142 GPQVIPAAFFASVAIF----FCFSAAALVAKQRSYLYLGSILGAALTYLSLASLVNIFLR 197
G ++ AA +V + + F AA K + +LG L AL L + +L+ IF
Sbjct 128 GKVILEAAILTTVVVLSLTVYTFWAAK---KGYDFNFLGPFLFGALIVLMVFALIQIFFP 184
Query 198 AQLVNNVILWGGL--FMYLGFVVYDTQLAVAQFDMGNRDYLLHALQFYVNFLSLFLRLVA 255
++ V+++G L ++ G++VYDT + ++ +Y+ A+ Y++ ++LFL L+
Sbjct 185 LGRIS-VMIYGCLAAIIFCGYIVYDTDNLIKRYSYD--EYIWAAVSLYLDIINLFLALLT 241
Query 256 I 256
I
Sbjct 242 I 242
> At4g02690
Length=248
Score = 35.4 bits (80), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 28/97 (28%), Positives = 51/97 (52%), Gaps = 5/97 (5%)
Query 168 KQRSYLYLGSILGAALTYLSLASLVNIFLRAQLVNNVILWGGL--FMYLGFVVYDTQLAV 225
K + +LG L ALT L +L+ I V+ V+++G L ++ G++VYDT +
Sbjct 155 KGYDFNFLGPFLFGALTVLIFFALIQILFPLGRVS-VMIYGCLVSIIFCGYIVYDTDNLI 213
Query 226 AQFDMGNRDYLLHALQFYVNFLSLFLRLVAILSERQE 262
+ +Y+ A+ Y++ ++LFL L+ +L Q
Sbjct 214 KRHTYD--EYIWAAVSLYLDIINLFLYLLTVLRALQR 248
> CE02441
Length=184
Score = 35.4 bits (80), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 22/81 (27%), Positives = 43/81 (53%), Gaps = 2/81 (2%)
Query 179 LGAALTYLSLASLVNIFLRAQLVNNVILWGGLFMYLGFVVYDTQLAVAQFDMGNRDYLLH 238
+G+ L L A + +F + VN VI G ++ +V D + + +F DY+
Sbjct 103 MGSLLCVLLWAGIFQMFFMSPAVNFVINVFGAGLFCVLLVIDLDMIMYRF--SPEDYICA 160
Query 239 ALQFYVNFLSLFLRLVAILSE 259
+ Y++ L+LF+R++ I++E
Sbjct 161 CVSLYMDILNLFIRILQIVAE 181
> 7299851
Length=264
Score = 35.0 bits (79), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 26/117 (22%), Positives = 55/117 (47%), Gaps = 6/117 (5%)
Query 147 PAAFFASVAIF--FCFSAAALVAKQR-SYLYLGSILGAALTYLSLASLVNIFLRAQLVNN 203
P F +V I C + R + +G +L + L L +V IF+ +V
Sbjct 144 PMEIFMAVLITASVCLGLTLFALQTRYDFTVMGGLLVSCLIILLFFGIVTIFVGGHMVTT 203
Query 204 VILWGGLFMYLGFVVYDTQLAVA---QFDMGNRDYLLHALQFYVNFLSLFLRLVAIL 257
+ ++ ++VYDTQL + ++ + +Y+ AL Y++ +++FL ++ ++
Sbjct 204 IYASLSALLFSVYLVYDTQLMMGGKHRYSISPEEYIFAALNIYMDVMNIFLDILQLI 260
> 7291631
Length=885
Score = 33.1 bits (74), Expect = 0.64, Method: Composition-based stats.
Identities = 14/34 (41%), Positives = 20/34 (58%), Gaps = 0/34 (0%)
Query 22 FKMEAFNFTGASAVPARSLLDFTPLSAPQKRHVS 55
FK + + A+ VP RSLL ++PQ RH+S
Sbjct 104 FKNKMLQYGAAAEVPVRSLLGHRSQASPQVRHIS 137
> SPBP23A10.08
Length=433
Score = 32.3 bits (72), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 15/39 (38%), Positives = 23/39 (58%), Gaps = 0/39 (0%)
Query 178 ILGAALTYLSLASLVNIFLRAQLVNNVILWGGLFMYLGF 216
ILG + + S ++ +RA L+NNVI+ GG + GF
Sbjct 324 ILGIPQLFQNCISECDVDIRASLLNNVIVCGGTSLMQGF 362
> Hs11545898
Length=311
Score = 31.6 bits (70), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 14/48 (29%), Positives = 27/48 (56%), Gaps = 2/48 (4%)
Query 216 FVVYDTQLAVA--QFDMGNRDYLLHALQFYVNFLSLFLRLVAILSERQ 261
F+ YDTQL + + + DY+ ALQ Y + + +F ++ ++ +R
Sbjct 264 FLAYDTQLVLGNRKHTISPEDYITGALQIYTDIIYIFTFVLQLMGDRN 311
> Hs4759118
Length=523
Score = 30.8 bits (68), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 26/78 (33%), Positives = 37/78 (47%), Gaps = 9/78 (11%)
Query 85 LSLMLSIGCVMGLTYSSQKAHAESQMLTKERAVYFGGFGVLNGMLAANYLHAV-HFYVGP 143
+S ++SI CV LT+S+ A S V GG V GM+AA++ V H YV
Sbjct 63 ISWIISI-CVFVLTFSAPLATVLSNRFGHRLVVMLGGLLVSTGMVAASFSQEVSHMYVAI 121
Query 144 QVIPAAFFASVAIFFCFS 161
+I + +CFS
Sbjct 122 GIIS-------GLGYCFS 132
> Hs13540555
Length=487
Score = 30.8 bits (68), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 21/74 (28%), Positives = 34/74 (45%), Gaps = 2/74 (2%)
Query 29 FTGASAVPARSLLDFTPLSAPQKRHVSRIY--AALTVNVLLTAVGVYGQLKWISLPPFLS 86
F V R + DF P K+ + + AA+T LL V +SLP ++S
Sbjct 354 FVNVGGVAMREIYDFMDDPKPHKKLGPQAWLVAAITATELLIVVKYDPHTLTLSLPFYIS 413
Query 87 LMLSIGCVMGLTYS 100
++G V+ LT++
Sbjct 414 QCWTLGSVLALTWT 427
> 7299350
Length=293
Score = 30.0 bits (66), Expect = 5.6, Method: Compositional matrix adjust.
Identities = 32/138 (23%), Positives = 57/138 (41%), Gaps = 26/138 (18%)
Query 50 QKRHVSRIY---------AALTVNVLLTAVGVYGQLKWISLPPFLSLMLSIGCVMGLTYS 100
+K+H+ +Y A + + VL +G+L S+P F+ L+L++GCV+ + Y
Sbjct 71 RKKHLGWVYLIHAVLSASAVIQLVVLRLCNKDFGELISPSVPSFVWLLLAVGCVLIMAYV 130
Query 101 SQKAHAESQMLTK----------------ERAVYFGGFGVLNGMLAAN-YLHAVHFYVGP 143
L R GVL+ +LA N L+ + Y+
Sbjct 131 YLANQCPCNGLLAIVIVEVIVIFVNCHRWARLSMLWMTGVLSLVLALNVMLYLMGVYLPL 190
Query 144 QVIPAAFFASVAIFFCFS 161
+++P + F V F C +
Sbjct 191 KILPGSIFMIVLTFCCIA 208
> 7290763
Length=1047
Score = 30.0 bits (66), Expect = 5.8, Method: Compositional matrix adjust.
Identities = 25/98 (25%), Positives = 48/98 (48%), Gaps = 10/98 (10%)
Query 167 AKQRSYLYLGSILGAALTYLSLASLVNIFLRAQLVNNVILWGGLFMYLGFVVYDTQLAVA 226
A Q L LG+ A L+ + + + L+A LV + GG F Y+ Q+A+A
Sbjct 339 ANQAQALTLGAPTTALLSMIVQSGCSQLLLKASLVRTLDRAGG-----AFAAYE-QVALA 392
Query 227 QFDMGNRDYLLHALQFYVNFLSLFLRLVAILSERQEEN 264
D D LLH +++ + L+ V ++++ ++++
Sbjct 393 SHD----DGLLHHQLLFIHLVRTMLQGVLVMTKEEDQH 426
> At1g66770
Length=261
Score = 30.0 bits (66), Expect = 6.1, Method: Compositional matrix adjust.
Identities = 21/66 (31%), Positives = 29/66 (43%), Gaps = 11/66 (16%)
Query 71 GVYGQLKWISLPPFLSLMLSIGCVMGLT--------YSSQKAHAESQMLTKERAVYFGGF 122
GV+ ++ PFL++ IGCV GL Y S K E + K R Y G
Sbjct 181 GVWTIYGFVPFDPFLAIPNGIGCVFGLVQLILYGTYYKSTKGIMEER---KNRLGYVGEV 237
Query 123 GVLNGM 128
G+ N +
Sbjct 238 GLSNAI 243
> YJL212c
Length=799
Score = 29.6 bits (65), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 21/82 (25%), Positives = 37/82 (45%), Gaps = 3/82 (3%)
Query 131 ANYLHAVHFYVGPQVIPAAFFASVAIFFCFSAAALVAKQRSYLYLGS---ILGAALTYLS 187
A ++H F+ GP IP + + ++FF S + ++R + ++GA +
Sbjct 688 AKHIHTPVFFTGPGNIPPSTPYNYSLFFAMSFCLNLIRKRWRAWFNKYNFVMGAGVEAGV 747
Query 188 LASLVNIFLRAQLVNNVILWGG 209
S+V IFL Q + W G
Sbjct 748 AISVVIIFLCVQYPGGKLSWWG 769
> 7292211
Length=1180
Score = 29.6 bits (65), Expect = 6.7, Method: Composition-based stats.
Identities = 22/77 (28%), Positives = 40/77 (51%), Gaps = 6/77 (7%)
Query 124 VLNGMLAANYLHAVHFYVGPQVIPAAFFASVAIFFCF---SAAALVAKQRSYLYLGSILG 180
+LN + A L+A++ ++ + F A+ FC+ + + AK + ++ SIL
Sbjct 155 ILNPIYDAYTLYAIYMFMPVPYLLQPFVLGSAVTFCYIINYSFVITAKDDNQMH--SILN 212
Query 181 AALTYLSLASLVNIFLR 197
A+ YLS +L+ IF R
Sbjct 213 EAI-YLSCVNLLGIFFR 228
> 7303389
Length=239
Score = 29.6 bits (65), Expect = 7.5, Method: Compositional matrix adjust.
Identities = 13/52 (25%), Positives = 32/52 (61%), Gaps = 3/52 (5%)
Query 209 GLFMYLGFVVYDTQLAVA---QFDMGNRDYLLHALQFYVNFLSLFLRLVAIL 257
G ++ ++VYDTQL + ++ + +Y+ AL Y++ +++F+ ++ I+
Sbjct 183 GALLFSVYLVYDTQLMLGGNHKYSISPEEYIFAALNLYLDIINIFMYILTII 234
> At5g06530
Length=567
Score = 29.3 bits (64), Expect = 8.1, Method: Compositional matrix adjust.
Identities = 37/130 (28%), Positives = 57/130 (43%), Gaps = 30/130 (23%)
Query 107 ESQMLTKERAV-------YFGGFGV----LNGMLAANYLHAVHFYVGPQVIPAAFFASV- 154
E ML KERA YF L+ +L + +L V+F G ++ P FF S+
Sbjct 451 ERAMLNKERAADMYRLSAYFLARTTSDLPLDFILPSLFLLVVYFMTGLRISPYPFFLSML 510
Query 155 AIFFCFSAAALVAKQRSYLYLGSILGAALTYLSLASLVNIFLRAQLVNNVILWGGLF--- 211
+F C ++A Q L +G+IL +LAS+ V +L GG F
Sbjct 511 TVFLC-----IIAAQGLGLAIGAILMDLKKATTLASVT--------VMTFMLAGGFFVKA 557
Query 212 --MYLGFVVY 219
++L F+ +
Sbjct 558 SPLFLDFLCF 567
Lambda K H
0.327 0.138 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5879395636
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40