bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0562_orf1
Length=94
Score E
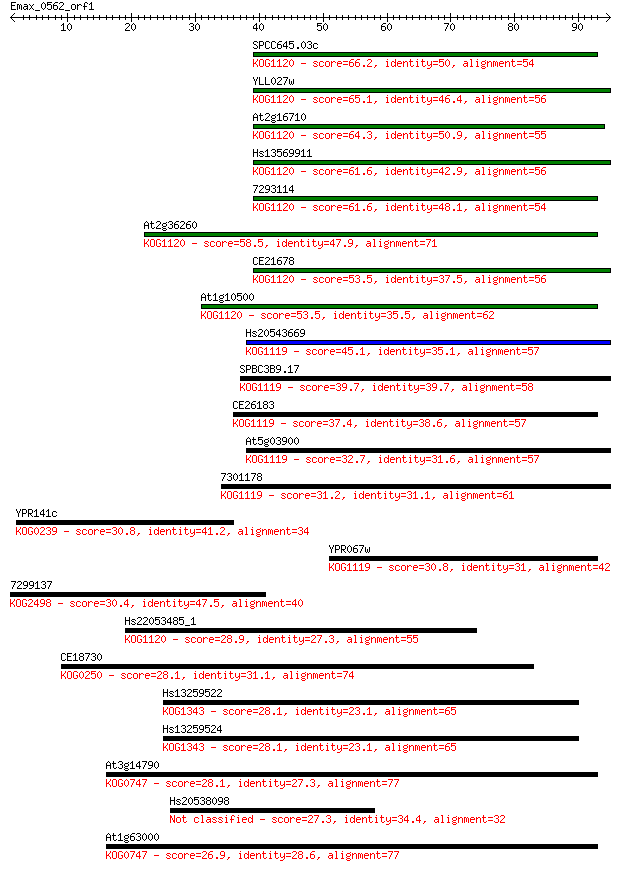
Sequences producing significant alignments: (Bits) Value
SPCC645.03c 66.2 1e-11
YLL027w 65.1 3e-11
At2g16710 64.3 5e-11
Hs13569911 61.6 3e-10
7293114 61.6 4e-10
At2g36260 58.5 3e-09
CE21678 53.5 8e-08
At1g10500 53.5 1e-07
Hs20543669 45.1 3e-05
SPBC3B9.17 39.7 0.001
CE26183 37.4 0.007
At5g03900 32.7 0.15
7301178 31.2 0.51
YPR141c 30.8 0.62
YPR067w 30.8 0.69
7299137 30.4 0.74
Hs22053485_1 28.9 2.3
CE18730 28.1 4.1
Hs13259522 28.1 4.3
Hs13259524 28.1 4.3
At3g14790 28.1 4.6
Hs20538098 27.3 7.5
At1g63000 26.9 9.8
> SPCC645.03c
Length=190
Score = 66.2 bits (160), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 27/54 (50%), Positives = 42/54 (77%), Gaps = 0/54 (0%)
Query 39 DEVVVVEGVEIRVAADAVMLLIGTQVDFVDEDVQTGFIFNNPNQKHSCGCGKSF 92
DE+V +G+ I VA A++ +IG+ +D+ D+D+Q+ FIF+NPN K +CGCG+SF
Sbjct 133 DEIVKQDGISIIVARRALLQIIGSVMDYRDDDLQSRFIFSNPNVKSTCGCGESF 186
> YLL027w
Length=250
Score = 65.1 bits (157), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 26/56 (46%), Positives = 43/56 (76%), Gaps = 0/56 (0%)
Query 39 DEVVVVEGVEIRVAADAVMLLIGTQVDFVDEDVQTGFIFNNPNQKHSCGCGKSFMV 94
DEVV +GV+I + + A+ +IG+++D++D+ + + F+F NPN K +CGCG+SFMV
Sbjct 195 DEVVEQDGVKIVIDSKALFSIIGSEMDWIDDKLASKFVFKNPNSKGTCGCGESFMV 250
> At2g16710
Length=137
Score = 64.3 bits (155), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 28/55 (50%), Positives = 41/55 (74%), Gaps = 0/55 (0%)
Query 39 DEVVVVEGVEIRVAADAVMLLIGTQVDFVDEDVQTGFIFNNPNQKHSCGCGKSFM 93
DE+V +GV I V A+M +IGT++DFVD+ +++ F+F NPN + CGCG+SFM
Sbjct 71 DELVEEKGVRILVEPKALMHVIGTKMDFVDDKLRSEFVFINPNSQGQCGCGESFM 125
> Hs13569911
Length=130
Score = 61.6 bits (148), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 24/56 (42%), Positives = 41/56 (73%), Gaps = 0/56 (0%)
Query 39 DEVVVVEGVEIRVAADAVMLLIGTQVDFVDEDVQTGFIFNNPNQKHSCGCGKSFMV 94
DE V+ +GV + + A + L+GT++D+V++ + + F+FNNPN K +CGCG+SF +
Sbjct 75 DEEVIQDGVRVFIEKKAQLTLLGTEMDYVEDKLSSEFVFNNPNIKGTCGCGESFNI 130
> 7293114
Length=131
Score = 61.6 bits (148), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 26/54 (48%), Positives = 40/54 (74%), Gaps = 0/54 (0%)
Query 39 DEVVVVEGVEIRVAADAVMLLIGTQVDFVDEDVQTGFIFNNPNQKHSCGCGKSF 92
DE VV +GV++ + A + L+GT++DFV+ + + F+FNNPN K +CGCG+SF
Sbjct 76 DEEVVQDGVKVFIDKKAQLSLLGTEMDFVESKLSSEFVFNNPNIKGTCGCGESF 129
> At2g36260
Length=109
Score = 58.5 bits (140), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 34/74 (45%), Positives = 48/74 (64%), Gaps = 4/74 (5%)
Query 22 ASGPLGLS---GHRRKNWGEDEVVVVEGVEIRVAADAVMLLIGTQVDFVDEDVQTGFIFN 78
A G GLS + ++ DEVV +GV+I V AVM +IGT++DFVD+ +++ F+F
Sbjct 34 AKGCNGLSYVLNYAQEKGKFDEVVEEKGVKILVDPKAVMHVIGTEMDFVDDKLRSEFVFV 93
Query 79 NPNQKHSCGCGKSF 92
NPN CGCG+SF
Sbjct 94 NPNAT-KCGCGESF 106
> CE21678
Length=129
Score = 53.5 bits (127), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 21/56 (37%), Positives = 39/56 (69%), Gaps = 0/56 (0%)
Query 39 DEVVVVEGVEIRVAADAVMLLIGTQVDFVDEDVQTGFIFNNPNQKHSCGCGKSFMV 94
DE V +G+++ + A + L+G+++D+V + + + F+F NPN K +CGCG+SF +
Sbjct 74 DEEVEQDGIKVWIEPKAQLSLLGSEMDYVTDKLSSEFVFRNPNIKGTCGCGESFSI 129
> At1g10500
Length=180
Score = 53.5 bits (127), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 22/62 (35%), Positives = 34/62 (54%), Gaps = 0/62 (0%)
Query 31 HRRKNWGEDEVVVVEGVEIRVAADAVMLLIGTQVDFVDEDVQTGFIFNNPNQKHSCGCGK 90
+R +D + +G I +++ L G Q+D+ D + GF F+NPN +CGCGK
Sbjct 115 NRANARPDDSTIEYQGFTIVCDPKSMLFLFGMQLDYSDALIGGGFSFSNPNATQTCGCGK 174
Query 91 SF 92
SF
Sbjct 175 SF 176
> Hs20543669
Length=154
Score = 45.1 bits (105), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 20/58 (34%), Positives = 32/58 (55%), Gaps = 1/58 (1%)
Query 38 EDEVVVVEGVEIRVAADAVMLLIGTQVDFVDEDVQTGF-IFNNPNQKHSCGCGKSFMV 94
+D V G + V +D++ + G QVDF E +++ F + NNP + C CG SF +
Sbjct 95 DDRVFEQGGARVVVDSDSLAFVKGAQVDFSQELIRSSFQVLNNPQAQQGCSCGSSFSI 152
> SPBC3B9.17
Length=205
Score = 39.7 bits (91), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 23/61 (37%), Positives = 36/61 (59%), Gaps = 4/61 (6%)
Query 37 GEDEVVVVEGVEIRVAADAVML--LIGTQVDFVDEDVQTGF-IFNNPNQKHSCGCGKSFM 93
G + V V G + RV AD + L + G+++++ +E + + F + NNP K SCGC SF
Sbjct 145 GNADTVFVRG-KARVVADNISLPLISGSEIEYTNELIGSSFQLLNNPRAKTSCGCNVSFD 203
Query 94 V 94
V
Sbjct 204 V 204
> CE26183
Length=203
Score = 37.4 bits (85), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 22/58 (37%), Positives = 30/58 (51%), Gaps = 5/58 (8%)
Query 36 WGEDEVVVVEGVEIRVAADAVMLLIGTQVDFVDEDVQTGF-IFNNPNQKHSCGCGKSF 92
W E G EI V ++ L G VDFV++ +++ F I NNP + C CG SF
Sbjct 145 WKSSE----NGAEIVVDELSLGFLKGATVDFVEDLMKSSFRIVNNPIAEKGCSCGSSF 198
> At5g03900
Length=757
Score = 32.7 bits (73), Expect = 0.15, Method: Composition-based stats.
Identities = 18/58 (31%), Positives = 30/58 (51%), Gaps = 1/58 (1%)
Query 38 EDEVVVVEGVEIRVAADAVMLLIGTQVDFVDEDVQTGFIFN-NPNQKHSCGCGKSFMV 94
+D V GV++ V + L+ G +D+ +E ++ F+ NP+ C C SFMV
Sbjct 698 DDRVFEKNGVKLVVDNVSYDLVKGATIDYEEELIRAAFVVAVNPSAVGGCSCKSSFMV 755
> 7301178
Length=120
Score = 31.2 bits (69), Expect = 0.51, Method: Compositional matrix adjust.
Identities = 19/63 (30%), Positives = 31/63 (49%), Gaps = 9/63 (14%)
Query 34 KNWGEDEV-VVVEGVEIRVAADAVMLLIGTQVDFVDEDVQTGF-IFNNPNQKHSCGCGKS 91
+ +GE E VV++ V + + G VD+ E ++ GF + NP + C CG S
Sbjct 63 RQFGEAEAKVVIDTVSLEYCS-------GATVDYHSELIRAGFRMVANPLAEQGCSCGSS 115
Query 92 FMV 94
F +
Sbjct 116 FSI 118
> YPR141c
Length=729
Score = 30.8 bits (68), Expect = 0.62, Method: Composition-based stats.
Identities = 14/35 (40%), Positives = 22/35 (62%), Gaps = 1/35 (2%)
Query 2 ERQPR-PHDGSATEASATPSHASGPLGLSGHRRKN 35
E PR P G +T+ +TPS + L ++GH+R+N
Sbjct 2 ESLPRTPTKGRSTQHLSTPSPKNDILAMNGHKRRN 36
> YPR067w
Length=185
Score = 30.8 bits (68), Expect = 0.69, Method: Compositional matrix adjust.
Identities = 13/42 (30%), Positives = 24/42 (57%), Gaps = 0/42 (0%)
Query 51 VAADAVMLLIGTQVDFVDEDVQTGFIFNNPNQKHSCGCGKSF 92
+ + ++ +L T + + +E + + F N + K SCGCG SF
Sbjct 140 IDSKSLNILNNTTLTYTNELIGSSFKIINGSLKSSCGCGSSF 181
> 7299137
Length=557
Score = 30.4 bits (67), Expect = 0.74, Method: Composition-based stats.
Identities = 19/49 (38%), Positives = 22/49 (44%), Gaps = 9/49 (18%)
Query 1 TERQPRPHDGS---------ATEASATPSHASGPLGLSGHRRKNWGEDE 40
T R P P G+ AT ATPS A G GLSG ++K E
Sbjct 25 TPRAPPPGSGTSSTSAGGSLATATFATPSTAPGTTGLSGEKKKGQSSSE 73
> Hs22053485_1
Length=114
Score = 28.9 bits (63), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 15/55 (27%), Positives = 30/55 (54%), Gaps = 4/55 (7%)
Query 19 PSHASGPLGLSGHRRKNWGEDEVVVVEGVEIRVAADAVMLLIGTQVDFVDEDVQT 73
P H +G+ K G+ V+ +GV + + A + L+GT++D+V+E + +
Sbjct 43 PEH----VGVKVEYTKTKGDSGEVIQDGVRVFIEKKAKLTLLGTEMDYVEEKLSS 93
> CE18730
Length=1130
Score = 28.1 bits (61), Expect = 4.1, Method: Composition-based stats.
Identities = 23/76 (30%), Positives = 34/76 (44%), Gaps = 3/76 (3%)
Query 9 DGSATEASATPSHASGPLGLSGHRRKNWGEDEVVVVEGVEIRVAAD--AVMLLIGTQVDF 66
DGS A+ S G GH R +G D+ V EG R+ D + + + TQ D
Sbjct 626 DGSQAYANGPNSQYRFYSGRGGHARGTFGNDQGDVDEGALARLIEDTKSEAMRLETQ-DL 684
Query 67 VDEDVQTGFIFNNPNQ 82
+D + I+N +Q
Sbjct 685 RKQDHELKVIYNERDQ 700
> Hs13259522
Length=952
Score = 28.1 bits (61), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 15/65 (23%), Positives = 24/65 (36%), Gaps = 0/65 (0%)
Query 25 PLGLSGHRRKNWGEDEVVVVEGVEIRVAADAVMLLIGTQVDFVDEDVQTGFIFNNPNQKH 84
PL GHR + + + A +L ++ TG I+++ KH
Sbjct 472 PLAQGGHRPLSRAQSSPAAPASLSAPEPASQARVLSSSETPARTLPFTTGLIYDSVMLKH 531
Query 85 SCGCG 89
C CG
Sbjct 532 QCSCG 536
> Hs13259524
Length=912
Score = 28.1 bits (61), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 15/65 (23%), Positives = 24/65 (36%), Gaps = 0/65 (0%)
Query 25 PLGLSGHRRKNWGEDEVVVVEGVEIRVAADAVMLLIGTQVDFVDEDVQTGFIFNNPNQKH 84
PL GHR + + + A +L ++ TG I+++ KH
Sbjct 432 PLAQGGHRPLSRAQSSPAAPASLSAPEPASQARVLSSSETPARTLPFTTGLIYDSVMLKH 491
Query 85 SCGCG 89
C CG
Sbjct 492 QCSCG 496
> At3g14790
Length=664
Score = 28.1 bits (61), Expect = 4.6, Method: Composition-based stats.
Identities = 21/77 (27%), Positives = 31/77 (40%), Gaps = 3/77 (3%)
Query 16 SATPSHASGPLGLSGHRRKNWGEDEVVVVEGVEIRVAADAVMLLIGTQVDFVDEDVQTGF 75
S PSH GL+G +W E E + + VA + + + D + + TG
Sbjct 425 SIKPSHVFNAAGLTGRPNVDWCESH--KTETIRVNVAGTLTLADVCRENDLLMMNFATGC 482
Query 76 IFNNPNQKHSCGCGKSF 92
IF + H G G F
Sbjct 483 IFEY-DAAHPEGSGIGF 498
> Hs20538098
Length=474
Score = 27.3 bits (59), Expect = 7.5, Method: Compositional matrix adjust.
Identities = 11/32 (34%), Positives = 16/32 (50%), Gaps = 0/32 (0%)
Query 26 LGLSGHRRKNWGEDEVVVVEGVEIRVAADAVM 57
G GHRR W E +V V E ++ +A +
Sbjct 341 WGAGGHRRHQWLEAQVEVASAKEFQIVFEATL 372
> At1g63000
Length=301
Score = 26.9 bits (58), Expect = 9.8, Method: Composition-based stats.
Identities = 22/77 (28%), Positives = 30/77 (38%), Gaps = 3/77 (3%)
Query 16 SATPSHASGPLGLSGHRRKNWGEDEVVVVEGVEIRVAADAVMLLIGTQVDFVDEDVQTGF 75
S PSH G++G +W E VE + VA + I + V + TG
Sbjct 58 SVKPSHVFNAAGVTGRPNVDWCESH--KVETIRTNVAGTLTLADICREKGLVLINYATGC 115
Query 76 IFNNPNQKHSCGCGKSF 92
IF + H G G F
Sbjct 116 IFEY-DSGHPLGSGIGF 131
Lambda K H
0.315 0.133 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1164659894
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40