bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0569_orf1
Length=127
Score E
Sequences producing significant alignments: (Bits) Value
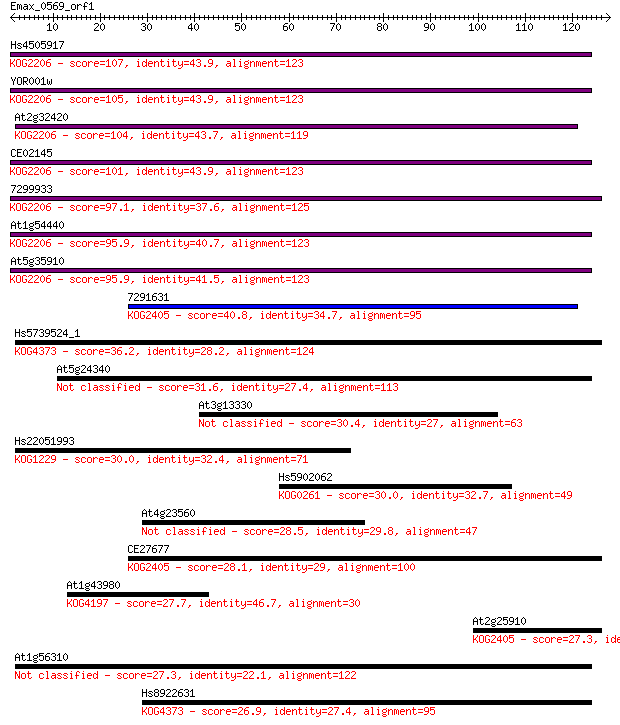
Hs4505917 107 7e-24
YOR001w 105 2e-23
At2g32420 104 4e-23
CE02145 101 4e-22
7299933 97.1 7e-21
At1g54440 95.9 1e-20
At5g35910 95.9 2e-20
7291631 40.8 6e-04
Hs5739524_1 36.2 0.014
At5g24340 31.6 0.37
At3g13330 30.4 0.80
Hs22051993 30.0 1.1
Hs5902062 30.0 1.2
At4g23560 28.5 2.8
CE27677 28.1 4.6
At1g43980 27.7 5.9
At2g25910 27.3 7.0
At1g56310 27.3 7.4
Hs8922631 26.9 8.2
> Hs4505917
Length=860
Score = 107 bits (266), Expect = 7e-24, Method: Composition-based stats.
Identities = 54/123 (43%), Positives = 76/123 (61%), Gaps = 1/123 (0%)
Query 1 CLIQLGTAEYVYVIDALKLFDHLHILNAITANPTIVKVFHNGDNDIAWLQRDFNVFVVNA 60
CL+Q+ T ++ID L+L ++ILN +P IVKVFH D+DI WLQ+DF ++VVN
Sbjct 327 CLMQISTRTEDFIIDTLELRSDMYILNESLTDPAIVKVFHGADSDIEWLQKDFGLYVVNM 386
Query 61 FDTGKAAKYLGVPGGTSLRNILQREFKVIKNERMSTCDWSRRPLTWDMRKYAVKDVGYLL 120
FDT +AA+ L + G SL ++L+ V N++ DW RPL +M YA D YLL
Sbjct 387 FDTHQAARLLNL-GRHSLDHLLKLYCNVDSNKQYQLADWRIRPLPEEMLSYARDDTHYLL 445
Query 121 CLY 123
+Y
Sbjct 446 YIY 448
> YOR001w
Length=733
Score = 105 bits (263), Expect = 2e-23, Method: Composition-based stats.
Identities = 54/123 (43%), Positives = 75/123 (60%), Gaps = 1/123 (0%)
Query 1 CLIQLGTAEYVYVIDALKLFDHLHILNAITANPTIVKVFHNGDNDIAWLQRDFNVFVVNA 60
CL+Q+ T E Y++D LKL ++LHILN + NP+IVKVFH DI WLQRD ++VV
Sbjct 252 CLMQISTRERDYLVDTLKLRENLHILNEVFTNPSIVKVFHGAFMDIIWLQRDLGLYVVGL 311
Query 61 FDTGKAAKYLGVPGGTSLRNILQREFKVIKNERMSTCDWSRRPLTWDMRKYAVKDVGYLL 120
FDT A+K +G+P SL +L+ +++ DW RPL+ M YA D +LL
Sbjct 312 FDTYHASKAIGLP-RHSLAYLLENFANFKTSKKYQLADWRIRPLSKPMTAYARADTHFLL 370
Query 121 CLY 123
+Y
Sbjct 371 NIY 373
> At2g32420
Length=237
Score = 104 bits (259), Expect = 4e-23, Method: Compositional matrix adjust.
Identities = 52/119 (43%), Positives = 74/119 (62%), Gaps = 1/119 (0%)
Query 2 LIQLGTAEYVYVIDALKLFDHLHILNAITANPTIVKVFHNGDNDIAWLQRDFNVFVVNAF 61
LIQ+ T E +++D + L D + IL + ++P I KVFH DND+ WLQRDF+++VVN F
Sbjct 95 LIQISTHEEDFLVDTIALHDVMSILRPVFSDPNICKVFHGADNDVIWLQRDFHIYVVNMF 154
Query 62 DTGKAAKYLGVPGGTSLRNILQREFKVIKNERMSTCDWSRRPLTWDMRKYAVKDVGYLL 120
DT KA + L P SL +L+ V N+ + DW +RPL+ +M +YA D YLL
Sbjct 155 DTAKACEVLSKP-QRSLAYLLETVCGVATNKLLQREDWRQRPLSEEMVRYARTDAHYLL 212
> CE02145
Length=876
Score = 101 bits (251), Expect = 4e-22, Method: Composition-based stats.
Identities = 54/123 (43%), Positives = 72/123 (58%), Gaps = 1/123 (0%)
Query 1 CLIQLGTAEYVYVIDALKLFDHLHILNAITANPTIVKVFHNGDNDIAWLQRDFNVFVVNA 60
CLIQ+ T + ++ID ++DH+ +LN ANP I+KVFH D+D+ WLQRD+ V VVN
Sbjct 317 CLIQISTRDEDFIIDPFPIWDHVGMLNEPFANPRILKVFHGSDSDVLWLQRDYGVHVVNL 376
Query 61 FDTGKAAKYLGVPGGTSLRNILQREFKVIKNERMSTCDWSRRPLTWDMRKYAVKDVGYLL 120
FDT A K L P SL + R V+ +++ DW RPL M YA +D YLL
Sbjct 377 FDTYVAMKKLKYP-KFSLAYLTLRFADVVLDKQYQLADWRARPLRNAMINYAREDTHYLL 435
Query 121 CLY 123
Y
Sbjct 436 YSY 438
> 7299933
Length=900
Score = 97.1 bits (240), Expect = 7e-21, Method: Composition-based stats.
Identities = 47/125 (37%), Positives = 72/125 (57%), Gaps = 1/125 (0%)
Query 1 CLIQLGTAEYVYVIDALKLFDHLHILNAITANPTIVKVFHNGDNDIAWLQRDFNVFVVNA 60
CL+Q+ T Y+ D L L D +HILN + +P +K+ H D DI WLQRD ++++VN
Sbjct 310 CLVQMSTRSKDYIFDTLILRDDMHILNLVLTDPKKLKILHGADLDIEWLQRDLSLYIVNM 369
Query 61 FDTGKAAKYLGVPGGTSLRNILQREFKVIKNERMSTCDWSRRPLTWDMRKYAVKDVGYLL 120
FDT +AAK L + SL +L+ + ++ + DW RPL + YA +D +L+
Sbjct 370 FDTHRAAKALNM-ARLSLAYLLKHYLDLDVDKSLQLADWRMRPLPQQLVDYARQDTHFLI 428
Query 121 CLYYR 125
+Y R
Sbjct 429 YVYGR 433
> At1g54440
Length=738
Score = 95.9 bits (237), Expect = 1e-20, Method: Composition-based stats.
Identities = 50/124 (40%), Positives = 71/124 (57%), Gaps = 2/124 (1%)
Query 1 CLIQLGTAEYVYVIDALKLFDHLH-ILNAITANPTIVKVFHNGDNDIAWLQRDFNVFVVN 59
CL+Q+ T Y++D KL+DH+ L + +P KV H D DI WLQRDF ++V N
Sbjct 267 CLMQISTRTEDYIVDIFKLWDHIGPYLRELFKDPKKKKVIHGADRDIIWLQRDFGIYVCN 326
Query 60 AFDTGKAAKYLGVPGGTSLRNILQREFKVIKNERMSTCDWSRRPLTWDMRKYAVKDVGYL 119
FDTG+A++ L + SL +L+ V N+ DW RPL M++YA +D YL
Sbjct 327 LFDTGQASRVLKLE-RNSLEFLLKHYCGVAANKEYQKADWRIRPLPDVMKRYAREDTHYL 385
Query 120 LCLY 123
L +Y
Sbjct 386 LYIY 389
> At5g35910
Length=838
Score = 95.9 bits (237), Expect = 2e-20, Method: Composition-based stats.
Identities = 51/124 (41%), Positives = 69/124 (55%), Gaps = 2/124 (1%)
Query 1 CLIQLGTAEYVYVIDALKLFDHLH-ILNAITANPTIVKVFHNGDNDIAWLQRDFNVFVVN 59
CL+Q+ T Y++D KL H+ L I +P KV H D DI WLQRDF ++V N
Sbjct 267 CLMQISTRTEDYIVDTFKLRIHIGPYLREIFKDPKKKKVMHGADRDIIWLQRDFGIYVCN 326
Query 60 AFDTGKAAKYLGVPGGTSLRNILQREFKVIKNERMSTCDWSRRPLTWDMRKYAVKDVGYL 119
FDTG+A++ L + SL +LQ V N+ DW RPL +M +YA +D YL
Sbjct 327 LFDTGQASRVLNLE-RNSLEFLLQHFCGVTANKEYQNADWRIRPLPEEMTRYAREDTHYL 385
Query 120 LCLY 123
L +Y
Sbjct 386 LYIY 389
> 7291631
Length=885
Score = 40.8 bits (94), Expect = 6e-04, Method: Composition-based stats.
Identities = 33/122 (27%), Positives = 49/122 (40%), Gaps = 37/122 (30%)
Query 26 LNAITANPTIVKVFHNGDNDIAWLQRDFNVFVVNAFDTGKA---------------AKYL 70
L + + ++KV H+ ND A L F + + N FDT A AKY+
Sbjct 602 LKTVLEHDQVIKVIHDCRNDAANLYLQFGILLRNVFDTQAAHAILQYQESGKQVYKAKYI 661
Query 71 GV------------PGGTSLRNILQREFKVIKNERMSTCDWSRRPLTWDMRKYAVKDVGY 118
+ P L+ I +R+ K W++RPLT +M YA DV
Sbjct 662 SLNSLCEQYNAPCNPIKDQLKQIYRRDQKF----------WAKRPLTREMMLYAAGDVLV 711
Query 119 LL 120
L+
Sbjct 712 LI 713
> Hs5739524_1
Length=532
Score = 36.2 bits (82), Expect = 0.014, Method: Composition-based stats.
Identities = 35/130 (26%), Positives = 63/130 (48%), Gaps = 9/130 (6%)
Query 2 LIQLGTAE---YVYVIDALKLFDHLHILNAITANPTIVKVFHNGDNDIAWLQRDFNVFVV 58
LIQL +E Y++ + ++ +F L + N + K + D L RDF++ +
Sbjct 99 LIQLCVSESKCYLFHVSSMSVFPQG--LKMLLENKAVKKAGVGIEGDQWKLLRDFDIKLK 156
Query 59 NAFD-TGKAAKYLGVPGGTSLRNILQREF--KVIKNERMSTCDWSRRPLTWDMRKYAVKD 115
N + T A K L SL ++++ +++K++ + +WS+ PLT D + YA D
Sbjct 157 NFVELTDVANKKLKCTETWSLNSLVKHLLGKQLLKDKSIRCSNWSKFPLTEDQKLYAATD 216
Query 116 VGYLLCLYYR 125
Y + YR
Sbjct 217 -AYAGFIIYR 225
> At5g24340
Length=505
Score = 31.6 bits (70), Expect = 0.37, Method: Composition-based stats.
Identities = 31/137 (22%), Positives = 59/137 (43%), Gaps = 30/137 (21%)
Query 11 VYVID--ALKLFDHLHILNAITANPTIVKVFHNGDNDIAWLQRDFNVFVVNAFDTG--KA 66
V++ID ++ L +LN + +P ++K+ D+ +L + F + + G +
Sbjct 68 VFLIDLSSIHLPSVWELLNDMFVSPDVLKLGFRFKQDLVYLS---STFTQHGCEGGFQEV 124
Query 67 AKYLGVPGGTSLRNILQRE--------------------FKVIKNERMSTCDWSRRPLTW 106
+YL + TS+ N LQ + + ++ + DWS RPLT
Sbjct 125 KQYLDI---TSIYNYLQHKRFGRKAPKDIKSLAAICKEMLDISLSKELQCSDWSYRPLTE 181
Query 107 DMRKYAVKDVGYLLCLY 123
+ + YA D LL ++
Sbjct 182 EQKLYAATDAHCLLQIF 198
> At3g13330
Length=1709
Score = 30.4 bits (67), Expect = 0.80, Method: Composition-based stats.
Identities = 17/70 (24%), Positives = 34/70 (48%), Gaps = 8/70 (11%)
Query 41 NGD--NDIAWLQRDFNVFVVNAFDTGKAAKYLGVPGG-----TSLRNILQREFKVIKNER 93
NGD +D+ W++ F+ F++++F +G+A+ L V G SL+ ++ ++
Sbjct 1443 NGDSLDDVKWMETLFH-FIISSFKSGRASYLLDVIAGFLYPVMSLQETSHKDLSILAKAA 1501
Query 94 MSTCDWSRRP 103
W P
Sbjct 1502 FELLKWRVFP 1511
> Hs22051993
Length=761
Score = 30.0 bits (66), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 23/74 (31%), Positives = 36/74 (48%), Gaps = 11/74 (14%)
Query 2 LIQLGTAEYVYVIDALKLFDHLHILNAITAN---PTIVKVFHNGDNDIAWLQRDFNVFVV 58
L +L EYV+ + + H H+ IT N P I ++ DN+ +W DFN+F +
Sbjct 389 LRRLSGNEYVFTKNVHQ--SHSHLAMPITINDVPPCISQLL---DNEESW---DFNIFEL 440
Query 59 NAFDTGKAAKYLGV 72
A + YLG+
Sbjct 441 EAITHKRPLVYLGL 454
> Hs5902062
Length=1391
Score = 30.0 bits (66), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 16/53 (30%), Positives = 25/53 (47%), Gaps = 4/53 (7%)
Query 58 VNAFDTGKAAKYLGVPGGTSLRNILQREFKVIKNERMSTC----DWSRRPLTW 106
+ A +TGK + G +L ++ +E VI++ S C D S PLT
Sbjct 728 IEALNTGKLQQQPGCTAEETLEALILKELSVIRDHAGSACLRELDKSNSPLTM 780
> At4g23560
Length=479
Score = 28.5 bits (62), Expect = 2.8, Method: Composition-based stats.
Identities = 14/47 (29%), Positives = 21/47 (44%), Gaps = 0/47 (0%)
Query 29 ITANPTIVKVFHNGDNDIAWLQRDFNVFVVNAFDTGKAAKYLGVPGG 75
+ A +V F+NG ND+A + D FV + + PGG
Sbjct 271 VGAQALLVSEFYNGANDLAKFKSDVESFVCAMMPGSSSQQIKPTPGG 317
> CE27677
Length=574
Score = 28.1 bits (61), Expect = 4.6, Method: Composition-based stats.
Identities = 29/116 (25%), Positives = 46/116 (39%), Gaps = 16/116 (13%)
Query 26 LNAITANPTIVKVFHNGDNDIAWLQRDFNVFVVNAFDTGKAAKYLG----VPGGTSLRNI 81
I + +VKV H+ + L + V + N FDT A L +R I
Sbjct 363 FKGILESEKVVKVIHDARRVASLLAHKYAVHMRNVFDTQVAHSLLQHEKFNKSLNEMRPI 422
Query 82 ----LQREF-------KVIKNERMSTC-DWSRRPLTWDMRKYAVKDVGYLLCLYYR 125
LQR + + +MS C +W RP+T + + V++ LL Y+
Sbjct 423 SFINLQRVYYPQSIMLSDVTPRKMSMCPNWGVRPITEEFQLTIVEEAHCLLSALYQ 478
> At1g43980
Length=633
Score = 27.7 bits (60), Expect = 5.9, Method: Compositional matrix adjust.
Identities = 14/30 (46%), Positives = 19/30 (63%), Gaps = 0/30 (0%)
Query 13 VIDALKLFDHLHILNAITANPTIVKVFHNG 42
VI+AL+LFD + N IT N + +F NG
Sbjct 55 VINALQLFDDIPDKNTITWNVCLKGLFKNG 84
> At2g25910
Length=282
Score = 27.3 bits (59), Expect = 7.0, Method: Compositional matrix adjust.
Identities = 12/27 (44%), Positives = 18/27 (66%), Gaps = 0/27 (0%)
Query 99 WSRRPLTWDMRKYAVKDVGYLLCLYYR 125
W+ RP+T M + A DV +LL LY++
Sbjct 116 WTYRPMTELMIRAAADDVRFLLYLYHK 142
> At1g56310
Length=582
Score = 27.3 bits (59), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 27/143 (18%), Positives = 58/143 (40%), Gaps = 22/143 (15%)
Query 2 LIQLGTAEYVYVIDALKLFDHL---------HILNAITANPTIVKVFHNGDNDIAW---L 49
++Q+G+ ++++D +KL++ HIL + + + D+ ++
Sbjct 402 IMQIGSDTKIFILDLIKLYNDASEILDNCLSHILQSKSTLKLVSLTEDYPDHKLSSGYNF 461
Query 50 QRDFNVFVVNAFDTGKAAKY---------LGVPGGTSLRNILQREFKVIKNERMSTCDWS 100
Q D ++ D +Y P G L + ++ V N+ DW
Sbjct 462 QCDIKQLALSYGDLKCFERYDMLLDIQNVFNEPFG-GLAGLTKKILGVSLNKTRRNSDWE 520
Query 101 RRPLTWDMRKYAVKDVGYLLCLY 123
+RPL+ + +YA D L+ ++
Sbjct 521 QRPLSQNQLEYAALDAAVLIHIF 543
> Hs8922631
Length=496
Score = 26.9 bits (58), Expect = 8.2, Method: Composition-based stats.
Identities = 26/104 (25%), Positives = 45/104 (43%), Gaps = 13/104 (12%)
Query 29 ITANPTIVKVFHNGDNDIAWLQRDFNVFVVNAFDTGKAAKYLGVP-------GGTSLRNI 81
I A+ TI+KV D + L +D+ + V D +YL + G SL+++
Sbjct 30 ILADGTILKVGVGCSEDASKLLQDYGLVVRGCLDL----RYLAMRQRNNLLCNGLSLKSL 85
Query 82 LQR--EFKVIKNERMSTCDWSRRPLTWDMRKYAVKDVGYLLCLY 123
+ F + K+ + +W LT D YA +D + L+
Sbjct 86 AETVLNFPLDKSLLLRCSNWDAETLTEDQVIYAARDAQISVALF 129
Lambda K H
0.327 0.141 0.451
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1176738752
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40