bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0588_orf1
Length=101
Score E
Sequences producing significant alignments: (Bits) Value
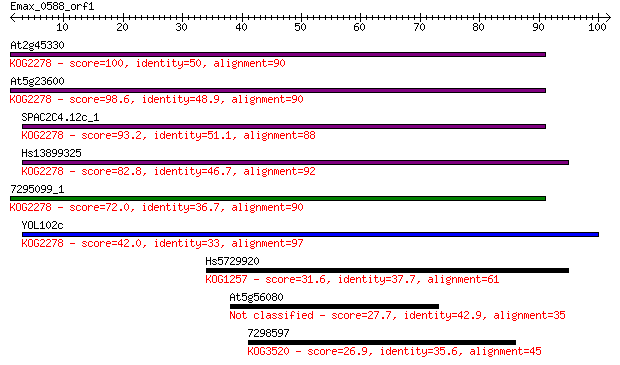
At2g45330 100 5e-22
At5g23600 98.6 3e-21
SPAC2C4.12c_1 93.2 1e-19
Hs13899325 82.8 1e-16
7295099_1 72.0 2e-13
YOL102c 42.0 3e-04
Hs5729920 31.6 0.33
At5g56080 27.7 5.4
7298597 26.9 8.3
> At2g45330
Length=257
Score = 100 bits (250), Expect = 5e-22, Method: Compositional matrix adjust.
Identities = 45/90 (50%), Positives = 64/90 (71%), Gaps = 0/90 (0%)
Query 1 VCVHGTYLWNWMSIRHLGLHRMHRNHIHFATGLPGEAAVLSGMRRNSEVAVYIHLEKAMK 60
VCVHGTY N SI GL RM+R H+HF+ GLP + V+SGMRRN V +++ ++KA++
Sbjct 157 VCVHGTYRKNLESILASGLKRMNRMHVHFSCGLPTDGEVISGMRRNVNVIIFLDIKKALE 216
Query 61 EGAIFFKSRNGVVLTEGLNGRLPPCFFSKV 90
+G F+ S N V+LTEG++G LP +F K+
Sbjct 217 DGIAFYISDNKVILTEGIDGVLPVDYFQKI 246
> At5g23600
Length=212
Score = 98.6 bits (244), Expect = 3e-21, Method: Compositional matrix adjust.
Identities = 44/90 (48%), Positives = 64/90 (71%), Gaps = 0/90 (0%)
Query 1 VCVHGTYLWNWMSIRHLGLHRMHRNHIHFATGLPGEAAVLSGMRRNSEVAVYIHLEKAMK 60
VCVHGTY N SI GL RM+R H+HF+ GLP + V+SG+RRN V +++H++KA++
Sbjct 112 VCVHGTYRKNLESILASGLKRMNRMHVHFSCGLPTDGEVISGVRRNVNVIIFLHIKKALE 171
Query 61 EGAIFFKSRNGVVLTEGLNGRLPPCFFSKV 90
+G F+ S N V+LT+G+ G LP +F K+
Sbjct 172 DGIAFYISDNKVILTQGIVGVLPVDYFQKI 201
> SPAC2C4.12c_1
Length=225
Score = 93.2 bits (230), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 45/88 (51%), Positives = 60/88 (68%), Gaps = 0/88 (0%)
Query 3 VHGTYLWNWMSIRHLGLHRMHRNHIHFATGLPGEAAVLSGMRRNSEVAVYIHLEKAMKEG 62
VHGT W I GL RM RNHIH ATGL G+ V+SG+R++ + +YI KAM++G
Sbjct 132 VHGTKKELWPVISKQGLSRMKRNHIHCATGLYGDPGVISGIRKSCTLYIYIDSAKAMQDG 191
Query 63 AIFFKSRNGVVLTEGLNGRLPPCFFSKV 90
F++S NGV+LTEG+NG L +FS+V
Sbjct 192 VEFYRSENGVILTEGVNGLLSSKYFSRV 219
> Hs13899325
Length=204
Score = 82.8 bits (203), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 43/93 (46%), Positives = 59/93 (63%), Gaps = 1/93 (1%)
Query 3 VHGTYLWNWMSIRHLGLHRMHRNHIHFATGLPGEAAVLSGMRRNSEVAVYIHLEKAMKEG 62
VHGT+ +W SI GL R HIH A GLPG+ ++SGMR + E+AV+I A+ +G
Sbjct 80 VHGTFWKHWPSILLKGLSCQGRTHIHLAPGLPGDPGIISGMRSHCEIAVFIDGPLALADG 139
Query 63 AIFFKSRNGVVLTEG-LNGRLPPCFFSKVVLLR 94
FF+S NGV+LT G +G L P +F + + LR
Sbjct 140 IPFFRSANGVILTPGNTDGFLLPKYFKEALQLR 172
> 7295099_1
Length=245
Score = 72.0 bits (175), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 33/90 (36%), Positives = 56/90 (62%), Gaps = 1/90 (1%)
Query 1 VCVHGTYLWNWMSIRHLGLHRMHRNHIHFATGLPGEAAVLSGMRRNSEVAVYIHLEKAMK 60
+ VHGTY +W +IR GL RM+RNH+HFA + LSG R + ++ +Y+++EK +
Sbjct 109 LAVHGTYYRHWGAIRSQGLSRMNRNHVHFACS-DETNSTLSGFRSDCQILIYLNVEKVLA 167
Query 61 EGAIFFKSRNGVVLTEGLNGRLPPCFFSKV 90
+G ++S N V+L G+ G + +F ++
Sbjct 168 DGIPIYRSSNNVLLCPGIEGFIHSSYFQQM 197
> YOL102c
Length=230
Score = 42.0 bits (97), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 32/102 (31%), Positives = 51/102 (50%), Gaps = 9/102 (8%)
Query 3 VHGTYLWNWMSIRHLG-LHRMHRNHIHFATGLPGEAAVLSGMRRNSEVAVYIHLEKAM-K 60
+HGT L + + I G + M RNH+H + G+ V+SGMR +S V ++I +
Sbjct 116 IHGTNLQSVIKIIESGAISPMSRNHVHLSPGMLHAKGVISGMRSSSNVYIFIDCHSPLFF 175
Query 61 EGAIFFKSRNGVVLTEGLNGRLPPCFFSKVVL---LRDGVEL 99
+ F+S N V L+ +P KVV+ L+D +L
Sbjct 176 QTLKMFRSLNNVYLSSS----IPVELIQKVVVKGNLKDEEKL 213
> Hs5729920
Length=604
Score = 31.6 bits (70), Expect = 0.33, Method: Composition-based stats.
Identities = 23/62 (37%), Positives = 31/62 (50%), Gaps = 8/62 (12%)
Query 34 PGEAAVLSGMRRNSEVAVYIHLEKAMKEGAIFFKSRNGVVLTEGLNGRLPPCFFSK-VVL 92
PG A + +R +V HL K M A + R L G++G +PPCF S+ V L
Sbjct 38 PGPARPVPLKKRGYDVTRNPHLNKGM---AFTLEER----LQLGIHGLIPPCFLSQDVQL 90
Query 93 LR 94
LR
Sbjct 91 LR 92
> At5g56080
Length=320
Score = 27.7 bits (60), Expect = 5.4, Method: Composition-based stats.
Identities = 15/36 (41%), Positives = 21/36 (58%), Gaps = 1/36 (2%)
Query 38 AVLSGMRRNSEVAVYIHLEKAMKEGA-IFFKSRNGV 72
A L GM + S+V HLEK M GA + +S +G+
Sbjct 202 AALVGMDKESKVKAIEHLEKHMAPGAVVMLRSAHGL 237
> 7298597
Length=1309
Score = 26.9 bits (58), Expect = 8.3, Method: Composition-based stats.
Identities = 16/46 (34%), Positives = 27/46 (58%), Gaps = 1/46 (2%)
Query 41 SGMRRNSEVAVYIHLEKAMKEGAIFFKSRNGVVLTEGL-NGRLPPC 85
S +R ++++ VY +EK K G+IFF+ + T+ L G+L C
Sbjct 298 SLLRAHAKIEVYDTVEKRPKRGSIFFRKKKPKAKTKSLVGGQLSAC 343
Lambda K H
0.328 0.141 0.453
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1184494980
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40