bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0597_orf1
Length=121
Score E
Sequences producing significant alignments: (Bits) Value
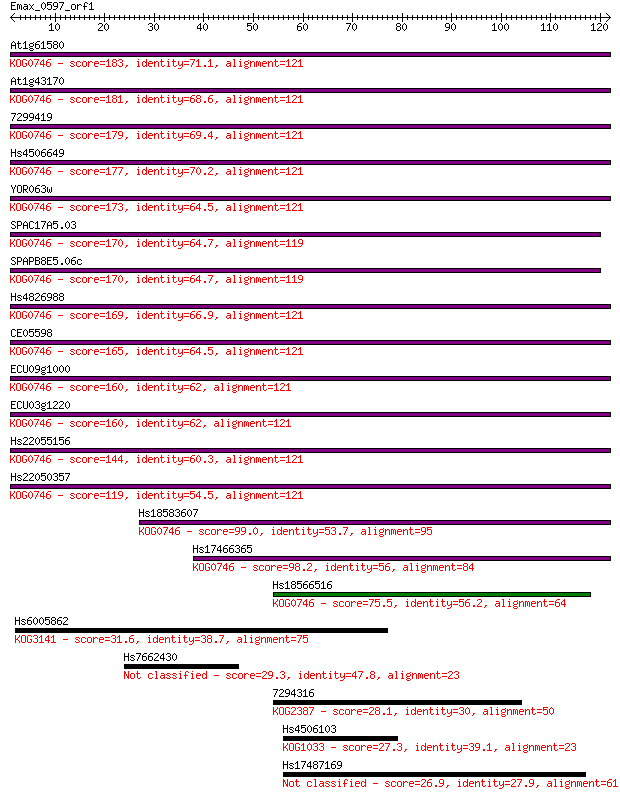
At1g61580 183 9e-47
At1g43170 181 4e-46
7299419 179 1e-45
Hs4506649 177 4e-45
YOR063w 173 6e-44
SPAC17A5.03 170 5e-43
SPAPB8E5.06c 170 5e-43
Hs4826988 169 2e-42
CE05598 165 2e-41
ECU09g1000 160 7e-40
ECU03g1220 160 7e-40
Hs22055156 144 4e-35
Hs22050357 119 1e-27
Hs18583607 99.0 2e-21
Hs17466365 98.2 3e-21
Hs18566516 75.5 2e-14
Hs6005862 31.6 0.38
Hs7662430 29.3 2.0
7294316 28.1 4.0
Hs4506103 27.3 7.3
Hs17487169 26.9 8.3
> At1g61580
Length=390
Score = 183 bits (464), Expect = 9e-47, Method: Compositional matrix adjust.
Identities = 86/122 (70%), Positives = 100/122 (81%), Gaps = 1/122 (0%)
Query 1 DEMLDVIGVTKGHGVKGVVSRFGVTRLPRKTHRGLRKVACIGAWHPARVQFQVPRHGQKG 60
DEM+D+IGVTKG G +GVV+R+GVTRLPRKTHRGLRKVACIGAWHPARV + V R GQ G
Sbjct 215 DEMIDIIGVTKGKGYEGVVTRWGVTRLPRKTHRGLRKVACIGAWHPARVSYTVARAGQNG 274
Query 61 YFHRTEMNKKVYRIGS-GNDPRNGSTDADLTEKRITPMGGFPHYGEVRSDFIMVKGCIVG 119
Y HRTEMNKKVYR+G G + + T+ D TEK ITPMGGFPHYG V+ D++M+KGC VG
Sbjct 275 YHHRTEMNKKVYRVGKVGQETHSAMTEYDRTEKDITPMGGFPHYGIVKEDYLMIKGCCVG 334
Query 120 SK 121
K
Sbjct 335 PK 336
> At1g43170
Length=389
Score = 181 bits (458), Expect = 4e-46, Method: Compositional matrix adjust.
Identities = 83/122 (68%), Positives = 99/122 (81%), Gaps = 1/122 (0%)
Query 1 DEMLDVIGVTKGHGVKGVVSRFGVTRLPRKTHRGLRKVACIGAWHPARVQFQVPRHGQKG 60
DEM+D+IGVTKG G +GVV+R+GVTRLPRKTHRGLRKVACIGAWHPARV + V R GQ G
Sbjct 215 DEMIDIIGVTKGKGYEGVVTRWGVTRLPRKTHRGLRKVACIGAWHPARVSYTVARAGQNG 274
Query 61 YFHRTEMNKKVYRIGS-GNDPRNGSTDADLTEKRITPMGGFPHYGEVRSDFIMVKGCIVG 119
Y HRTE+NKK+YR+G G + T+ D TEK +TPMGGFPHYG V+ D++M+KGC VG
Sbjct 275 YHHRTELNKKIYRLGKVGTEAHTAMTEYDRTEKDVTPMGGFPHYGIVKDDYLMIKGCCVG 334
Query 120 SK 121
K
Sbjct 335 PK 336
> 7299419
Length=403
Score = 179 bits (453), Expect = 1e-45, Method: Compositional matrix adjust.
Identities = 84/128 (65%), Positives = 100/128 (78%), Gaps = 7/128 (5%)
Query 1 DEMLDVIGVTKGHGVKGVVSRFGVTRLPRKTHRGLRKVACIGAWHPARVQFQVPRHGQKG 60
DEM+D +GVTKG G KGV SR+ +LPRKTH+GLRKVACIGAWHP+RV V R GQKG
Sbjct 201 DEMIDCVGVTKGKGFKGVTSRWHTKKLPRKTHKGLRKVACIGAWHPSRVSTTVARAGQKG 260
Query 61 YFHRTEMNKKVYRIGSGNDPR-------NGSTDADLTEKRITPMGGFPHYGEVRSDFIMV 113
Y HRTE+NKK+YRIG+G + N ST+ DLT+K ITPMGGFPHYGEV +DF+M+
Sbjct 261 YHHRTEINKKIYRIGAGIHTKDGKVIKNNASTEYDLTDKSITPMGGFPHYGEVNNDFVMI 320
Query 114 KGCIVGSK 121
KGC +GSK
Sbjct 321 KGCCIGSK 328
> Hs4506649
Length=403
Score = 177 bits (449), Expect = 4e-45, Method: Compositional matrix adjust.
Identities = 85/128 (66%), Positives = 100/128 (78%), Gaps = 7/128 (5%)
Query 1 DEMLDVIGVTKGHGVKGVVSRFGVTRLPRKTHRGLRKVACIGAWHPARVQFQVPRHGQKG 60
DEM+DVIGVTKG G KGV SR+ +LPRKTHRGLRKVACIGAWHPARV F V R GQKG
Sbjct 214 DEMIDVIGVTKGKGYKGVTSRWHTKKLPRKTHRGLRKVACIGAWHPARVAFSVARAGQKG 273
Query 61 YFHRTEMNKKVYRIGSGNDPR-------NGSTDADLTEKRITPMGGFPHYGEVRSDFIMV 113
Y HRTE+NKK+Y+IG G + N STD DL++K I P+GGF HYGEV +DF+M+
Sbjct 274 YHHRTEINKKIYKIGQGYLIKDGKLIKNNASTDYDLSDKSINPLGGFVHYGEVTNDFVML 333
Query 114 KGCIVGSK 121
KGC+VG+K
Sbjct 334 KGCVVGTK 341
> YOR063w
Length=387
Score = 173 bits (439), Expect = 6e-44, Method: Compositional matrix adjust.
Identities = 78/121 (64%), Positives = 96/121 (79%), Gaps = 0/121 (0%)
Query 1 DEMLDVIGVTKGHGVKGVVSRFGVTRLPRKTHRGLRKVACIGAWHPARVQFQVPRHGQKG 60
+EM+D I VTKGHG +GV R+G +LPRKTHRGLRKVACIGAWHPA V + V R GQ+G
Sbjct 212 NEMIDAIAVTKGHGFEGVTHRWGTKKLPRKTHRGLRKVACIGAWHPAHVMWSVARAGQRG 271
Query 61 YFHRTEMNKKVYRIGSGNDPRNGSTDADLTEKRITPMGGFPHYGEVRSDFIMVKGCIVGS 120
Y RT +N K+YR+G G+D NG+T D T+K ITPMGGF HYGE+++DFIMVKGCI G+
Sbjct 272 YHSRTSINHKIYRVGKGDDEANGATSFDRTKKTITPMGGFVHYGEIKNDFIMVKGCIPGN 331
Query 121 K 121
+
Sbjct 332 R 332
> SPAC17A5.03
Length=388
Score = 170 bits (431), Expect = 5e-43, Method: Compositional matrix adjust.
Identities = 77/119 (64%), Positives = 93/119 (78%), Gaps = 0/119 (0%)
Query 1 DEMLDVIGVTKGHGVKGVVSRFGVTRLPRKTHRGLRKVACIGAWHPARVQFQVPRHGQKG 60
+EM+DVIGVT+G G +G +R+G RLPRKTHRGLRKVACIGAWHPA VQ+ V R G G
Sbjct 212 NEMIDVIGVTRGKGNEGTTARWGTKRLPRKTHRGLRKVACIGAWHPANVQWTVARAGNAG 271
Query 61 YFHRTEMNKKVYRIGSGNDPRNGSTDADLTEKRITPMGGFPHYGEVRSDFIMVKGCIVG 119
Y HRT++N K+YRIG+G+D +N STD D TEKRITPMGGF YG V +DF+M+ G G
Sbjct 272 YMHRTQLNSKIYRIGAGDDAKNASTDFDATEKRITPMGGFVRYGVVENDFVMLNGATPG 330
> SPAPB8E5.06c
Length=388
Score = 170 bits (431), Expect = 5e-43, Method: Compositional matrix adjust.
Identities = 77/119 (64%), Positives = 93/119 (78%), Gaps = 0/119 (0%)
Query 1 DEMLDVIGVTKGHGVKGVVSRFGVTRLPRKTHRGLRKVACIGAWHPARVQFQVPRHGQKG 60
+EM+DVIGVT+G G +G +R+G RLPRKTHRGLRKVACIGAWHPA VQ+ V R G G
Sbjct 212 NEMIDVIGVTRGKGNEGTTARWGTKRLPRKTHRGLRKVACIGAWHPANVQWTVARAGNAG 271
Query 61 YFHRTEMNKKVYRIGSGNDPRNGSTDADLTEKRITPMGGFPHYGEVRSDFIMVKGCIVG 119
Y HRT++N K+YRIG+G+D +N STD D TEKRITPMGGF YG V +DF+M+ G G
Sbjct 272 YMHRTQLNSKIYRIGAGDDAKNASTDFDATEKRITPMGGFVRYGVVENDFVMLNGATPG 330
> Hs4826988
Length=407
Score = 169 bits (427), Expect = 2e-42, Method: Compositional matrix adjust.
Identities = 81/128 (63%), Positives = 96/128 (75%), Gaps = 7/128 (5%)
Query 1 DEMLDVIGVTKGHGVKGVVSRFGVTRLPRKTHRGLRKVACIGAWHPARVQFQVPRHGQKG 60
E++DVI VTKG GVKGV SR+ +LPRKTH+GLRKVACIGAWHPARV + R GQKG
Sbjct 214 SEVIDVIAVTKGRGVKGVTSRWHTKKLPRKTHKGLRKVACIGAWHPARVGCSIARAGQKG 273
Query 61 YFHRTEMNKKVYRIGSGND-------PRNGSTDADLTEKRITPMGGFPHYGEVRSDFIMV 113
Y HRTE+NKK++RIG G N ST D+T K ITP+GGFPHYGEV +DF+M+
Sbjct 274 YHHRTELNKKIFRIGRGPHMEDGKLVKNNASTSYDVTAKSITPLGGFPHYGEVNNDFVML 333
Query 114 KGCIVGSK 121
KGCI G+K
Sbjct 334 KGCIAGTK 341
> CE05598
Length=401
Score = 165 bits (418), Expect = 2e-41, Method: Compositional matrix adjust.
Identities = 78/125 (62%), Positives = 95/125 (76%), Gaps = 4/125 (3%)
Query 1 DEMLDVIGVTKGHGVKGVVSRFGVTRLPRKTHRGLRKVACIGAWHPARVQFQVPRHGQKG 60
DEM+D IGVT+GHG KGV SR+ +LPRKTH+GLRKVACIGAWHP+RV F V R GQKG
Sbjct 214 DEMIDTIGVTRGHGFKGVTSRWHTKKLPRKTHKGLRKVACIGAWHPSRVAFTVARAGQKG 273
Query 61 YFHRTEMNKKVYRIG----SGNDPRNGSTDADLTEKRITPMGGFPHYGEVRSDFIMVKGC 116
+ HRT +N K+YRIG + NGST+ DLT+K ITPMGGFP YG V D+IM++G
Sbjct 274 FHHRTIINNKIYRIGKSALTEEGKNNGSTEFDLTQKTITPMGGFPRYGIVNQDYIMLRGA 333
Query 117 IVGSK 121
++G K
Sbjct 334 VLGPK 338
> ECU09g1000
Length=383
Score = 160 bits (404), Expect = 7e-40, Method: Compositional matrix adjust.
Identities = 75/121 (61%), Positives = 95/121 (78%), Gaps = 2/121 (1%)
Query 1 DEMLDVIGVTKGHGVKGVVSRFGVTRLPRKTHRGLRKVACIGAWHPARVQFQVPRHGQKG 60
+E +D IGVTKG G +G V RFGV + PRK+ +G+RKVACIGAWHP+RV + + R GQ G
Sbjct 208 NENIDTIGVTKGKGFQGTVKRFGVRKQPRKSRKGIRKVACIGAWHPSRVMYSIARAGQMG 267
Query 61 YFHRTEMNKKVYRIGSGNDPRNGSTDADLTEKRITPMGGFPHYGEVRSDFIMVKGCIVGS 120
+ RTE NK+VY IG+G+ N T+ DLTEK I+PMGGFPHYGEVR+DFIMVKG +VG+
Sbjct 268 FHRRTEKNKRVYMIGNGSS--NIKTEFDLTEKPISPMGGFPHYGEVRNDFIMVKGAVVGA 325
Query 121 K 121
+
Sbjct 326 R 326
> ECU03g1220
Length=383
Score = 160 bits (404), Expect = 7e-40, Method: Compositional matrix adjust.
Identities = 75/121 (61%), Positives = 95/121 (78%), Gaps = 2/121 (1%)
Query 1 DEMLDVIGVTKGHGVKGVVSRFGVTRLPRKTHRGLRKVACIGAWHPARVQFQVPRHGQKG 60
+E +D IGVTKG G +G V RFGV + PRK+ +G+RKVACIGAWHP+RV + + R GQ G
Sbjct 208 NENIDTIGVTKGKGFQGTVKRFGVRKQPRKSRKGIRKVACIGAWHPSRVMYSIARAGQMG 267
Query 61 YFHRTEMNKKVYRIGSGNDPRNGSTDADLTEKRITPMGGFPHYGEVRSDFIMVKGCIVGS 120
+ RTE NK+VY IG+G+ N T+ DLTEK I+PMGGFPHYGEVR+DFIMVKG +VG+
Sbjct 268 FHRRTEKNKRVYMIGNGSS--NIKTEFDLTEKPISPMGGFPHYGEVRNDFIMVKGAVVGA 325
Query 121 K 121
+
Sbjct 326 R 326
> Hs22055156
Length=222
Score = 144 bits (363), Expect = 4e-35, Method: Compositional matrix adjust.
Identities = 73/128 (57%), Positives = 88/128 (68%), Gaps = 7/128 (5%)
Query 1 DEMLDVIGVTKGHGVKGVVSRFGVTRLPRKTHRGLRKVACIGAWHPARVQFQVPRHGQKG 60
DEM+ VIGVTKG G KGV S + T+LP KTHRGL KVACIGAWH A V F V GQKG
Sbjct 34 DEMIKVIGVTKGKGYKGVTSCWHTTKLPHKTHRGLCKVACIGAWHLAHVAFSVSHAGQKG 93
Query 61 YFHRTEMNKKVYRIGSGND-------PRNGSTDADLTEKRITPMGGFPHYGEVRSDFIMV 113
Y H TE+NKK+Y+IG G N S D DL++K I +GGF H G+V +DF+M+
Sbjct 94 YHHGTEINKKIYKIGQGYPIMDGKLIKNNASIDYDLSDKSINSLGGFVHCGKVTNDFVML 153
Query 114 KGCIVGSK 121
KG +VG+K
Sbjct 154 KGYVVGTK 161
> Hs22050357
Length=432
Score = 119 bits (299), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 66/128 (51%), Positives = 81/128 (63%), Gaps = 13/128 (10%)
Query 1 DEMLDVIGVTKGHGVKGVVSRFGVTRLPRKTHRGLRKVACIGAWHPARVQFQVPRHGQKG 60
D+M+DVI +TKG+G KGV SR+ +LP KTHRGL KVAC G HPA V V QKG
Sbjct 249 DKMIDVIRLTKGNGYKGVTSRWHTKKLPHKTHRGLCKVACTGTQHPAPVALPVACIWQKG 308
Query 61 YFHRTEMNKKVYRIGSGNDPR-------NGSTDADLTEKRITPMGGFPHYGEVRSDFIMV 113
Y H TE+N K+Y+I G R N STD DL P+G F HYGEV +DF+M+
Sbjct 309 YCHSTEIN-KIYKISQGYFVRDSKLIKNNASTDYDL-----HPLGAFVHYGEVTNDFVML 362
Query 114 KGCIVGSK 121
K C+V +K
Sbjct 363 KDCVVDTK 370
> Hs18583607
Length=169
Score = 99.0 bits (245), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 51/103 (49%), Positives = 68/103 (66%), Gaps = 9/103 (8%)
Query 27 LPRKTHRG-LRKVACIGAWHPARVQFQVPRHGQKGYFHRTEMNKKVYRIGSGNDPR---- 81
LPR+ G + + H A ++F V R GQKGY HRTE+NKK+Y+IG G +
Sbjct 6 LPRQEAEGPMSGSGELSVRHHA-IKFSVARAGQKGYHHRTEINKKIYKIGQGYLIKDGKL 64
Query 82 ---NGSTDADLTEKRITPMGGFPHYGEVRSDFIMVKGCIVGSK 121
N STD DL++K I P+GGF HYGEV +DF+M+KGC+VG+K
Sbjct 65 IKNNASTDYDLSDKSINPLGGFVHYGEVTNDFVMLKGCVVGTK 107
> Hs17466365
Length=251
Score = 98.2 bits (243), Expect = 3e-21, Method: Compositional matrix adjust.
Identities = 47/91 (51%), Positives = 60/91 (65%), Gaps = 7/91 (7%)
Query 38 VACIGAWHPARVQFQVPRHGQKGYFHRTEMNKKVYRIGSGNDPR-------NGSTDADLT 90
+ IGAWHP RV F R GQK Y H T++NKK+Y+IG + N STD DL+
Sbjct 146 IDVIGAWHPDRVAFSGARTGQKEYHHCTKINKKIYKIGQSYLIKGGKLIKNNASTDYDLS 205
Query 91 EKRITPMGGFPHYGEVRSDFIMVKGCIVGSK 121
K I P+GGF HYGEV +DF+M+KGC+V +K
Sbjct 206 VKSINPLGGFVHYGEVTNDFVMLKGCVVVTK 236
> Hs18566516
Length=364
Score = 75.5 bits (184), Expect = 2e-14, Method: Composition-based stats.
Identities = 36/71 (50%), Positives = 46/71 (64%), Gaps = 7/71 (9%)
Query 54 PRHGQKGYFHRTEMNKKVYRIGSG---NDPR----NGSTDADLTEKRITPMGGFPHYGEV 106
P QKGY H TE NKK+ IG G D + N STD DL++K I P+GGF HY E+
Sbjct 261 PMEIQKGYHHCTENNKKICNIGHGYLTKDEKWIKNNASTDGDLSDKSINPLGGFVHYNEM 320
Query 107 RSDFIMVKGCI 117
+DF+M+KGC+
Sbjct 321 NNDFVMLKGCV 331
> Hs6005862
Length=348
Score = 31.6 bits (70), Expect = 0.38, Method: Compositional matrix adjust.
Identities = 29/82 (35%), Positives = 38/82 (46%), Gaps = 12/82 (14%)
Query 2 EMLDVIGVTKGHGVKGVVSRFGVTRLP-----RKTHRGLRKVAC--IGAWHPARVQFQVP 54
+ +DV T G G +GV+ R+G P KTHR VA IG RV
Sbjct 202 QYVDVTAKTIGKGFQGVMKRWGFKGQPATHGQTKTHRRPGAVATGDIG-----RVWPGTK 256
Query 55 RHGQKGYFHRTEMNKKVYRIGS 76
G+ G +RTE KV+RI +
Sbjct 257 MPGKMGNIYRTEYGLKVWRINT 278
> Hs7662430
Length=494
Score = 29.3 bits (64), Expect = 2.0, Method: Composition-based stats.
Identities = 11/26 (42%), Positives = 16/26 (61%), Gaps = 3/26 (11%)
Query 24 VTRLPRKTHRGLRKVACIGAW---HP 46
+ RLPR + +G R++ C AW HP
Sbjct 442 LARLPRSSRQGHRRMVCCSAWSEDHP 467
> 7294316
Length=627
Score = 28.1 bits (61), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 15/50 (30%), Positives = 23/50 (46%), Gaps = 0/50 (0%)
Query 54 PRHGQKGYFHRTEMNKKVYRIGSGNDPRNGSTDADLTEKRITPMGGFPHY 103
P+ Q+ + HR E+N K + R TD D T I + G P++
Sbjct 471 PKSVQERHRHRYEVNPKYVHLLEEQGMRFVGTDVDKTRMEIIELSGHPYF 520
> Hs4506103
Length=551
Score = 27.3 bits (59), Expect = 7.3, Method: Composition-based stats.
Identities = 9/23 (39%), Positives = 17/23 (73%), Gaps = 0/23 (0%)
Query 56 HGQKGYFHRTEMNKKVYRIGSGN 78
HG +G+ ++ +M +K Y IG+G+
Sbjct 126 HGPEGFHYKCKMGQKEYSIGTGS 148
> Hs17487169
Length=276
Score = 26.9 bits (58), Expect = 8.3, Method: Compositional matrix adjust.
Identities = 17/69 (24%), Positives = 32/69 (46%), Gaps = 10/69 (14%)
Query 56 HGQKGYFHRTEMNKKVYRIGSGNDPRNGSTDA--------DLTEKRITPMGGFPHYGEVR 107
H +G HR E++K + G+ DP +G+T + +L++ + P G E +
Sbjct 57 HSSRGRTHRPELSKHSLQPGTRADPSSGTTHSSRGRMHRPELSKHSLQP--GMHTQTEAQ 114
Query 108 SDFIMVKGC 116
S ++ G
Sbjct 115 SKHLLQPGT 123
Lambda K H
0.322 0.141 0.442
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1198419392
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40