bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0600_orf1
Length=139
Score E
Sequences producing significant alignments: (Bits) Value
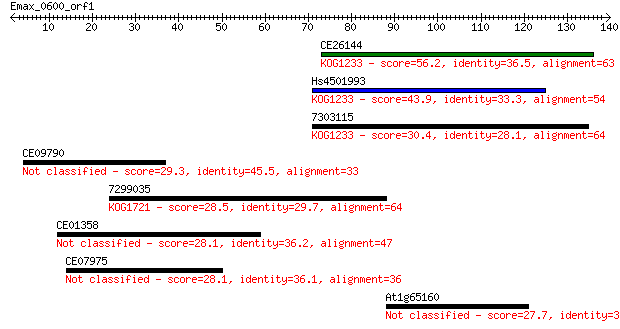
CE26144 56.2 2e-08
Hs4501993 43.9 8e-05
7303115 30.4 1.1
CE09790 29.3 2.2
7299035 28.5 4.0
CE01358 28.1 5.1
CE07975 28.1 5.8
At1g65160 27.7 7.2
> CE26144
Length=597
Score = 56.2 bits (134), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 23/63 (36%), Positives = 38/63 (60%), Gaps = 0/63 (0%)
Query 73 RVKMCKWNGWGYADTYVSMSSSSMIRLHGNRYSLCGVDMGHWHEFMESIPGIEFTFTSPA 132
R K+ KWNGWGY+D+ +++ + G++Y + G M H+ + E+ GI+ F SPA
Sbjct 17 RDKILKWNGWGYSDSQFAINKDGHVTFTGDKYEISGKVMPHFRPWFENYLGIDLGFVSPA 76
Query 133 QNI 135
Q +
Sbjct 77 QKL 79
> Hs4501993
Length=658
Score = 43.9 bits (102), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 18/54 (33%), Positives = 31/54 (57%), Gaps = 0/54 (0%)
Query 71 RNRVKMCKWNGWGYADTYVSMSSSSMIRLHGNRYSLCGVDMGHWHEFMESIPGI 124
+ R ++ KWNGWGY D+ + I L G RY L G+ + + E++++ G+
Sbjct 85 KKRQEVMKWNGWGYNDSKFIFNKKGQIELTGKRYPLSGMGLPTFKEWIQNTLGV 138
> 7303115
Length=631
Score = 30.4 bits (67), Expect = 1.1, Method: Composition-based stats.
Identities = 18/75 (24%), Positives = 32/75 (42%), Gaps = 12/75 (16%)
Query 71 RNRVKMCKWNGWGYADTYVSMSSSSMIRLHGNRYSLCGVDMGHWHEFME----------- 119
+ R + KW GWGY D+ +I G +Y L G ++ + +++E
Sbjct 40 KKRHEALKWFGWGYNDSQF-YGKDGIICFRGEKYPLGGCELPSFTKWVEKKFDLRVDTTK 98
Query 120 SIPGIEFTFTSPAQN 134
P + T+ P +N
Sbjct 99 QYPQLPRTYPRPVEN 113
> CE09790
Length=1153
Score = 29.3 bits (64), Expect = 2.2, Method: Composition-based stats.
Identities = 15/33 (45%), Positives = 20/33 (60%), Gaps = 0/33 (0%)
Query 4 GEEEIPIEEVKKSNDVREYADPSEDNKGDASWI 36
G E IEEV D+R+ A PS+ NK +A+ I
Sbjct 1027 GLNEDEIEEVPSKTDLRKEAAPSKTNKRNANLI 1059
> 7299035
Length=323
Score = 28.5 bits (62), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 19/66 (28%), Positives = 31/66 (46%), Gaps = 2/66 (3%)
Query 24 DPSEDNKGDASWICPYMNSPSSLRLSLHVFPKKIHAGHSVIDCRLFGRNRVKM-C-KWNG 81
DP + G + W C + + L L +++HA ++IDCR R K+ C + +
Sbjct 103 DPENEPIGVSRWNCQHCGAGFQLSEVLRRHIQEVHASITIIDCRERRRIFTKLGCYQVHN 162
Query 82 WGYADT 87
YA T
Sbjct 163 CSYAKT 168
> CE01358
Length=558
Score = 28.1 bits (61), Expect = 5.1, Method: Composition-based stats.
Identities = 17/52 (32%), Positives = 28/52 (53%), Gaps = 5/52 (9%)
Query 12 EVKKSN-DVREYADPSEDNKGDASWICPYMNSPSS----LRLSLHVFPKKIH 58
EV +SN + EY DPS +GD + + P + +PS+ +L + + IH
Sbjct 437 EVNRSNPHLIEYPDPSSILRGDNAGLPPVIMAPSTNDEVCKLQIEALQRHIH 488
> CE07975
Length=3036
Score = 28.1 bits (61), Expect = 5.8, Method: Compositional matrix adjust.
Identities = 13/38 (34%), Positives = 20/38 (52%), Gaps = 2/38 (5%)
Query 14 KKSNDVREYADPSEDNKGDAS--WICPYMNSPSSLRLS 49
+++ND + P D G A W CP ++P +RLS
Sbjct 182 RRNNDTYPFVMPKPDCHGAAKNLWDCPAFSNPQKIRLS 219
> At1g65160
Length=309
Score = 27.7 bits (60), Expect = 7.2, Method: Compositional matrix adjust.
Identities = 11/33 (33%), Positives = 16/33 (48%), Gaps = 0/33 (0%)
Query 88 YVSMSSSSMIRLHGNRYSLCGVDMGHWHEFMES 120
Y SM+ LHGNRY+ + W ++ S
Sbjct 224 YTKYRLVSMVCLHGNRYNCVAYENNRWVRYLRS 256
Lambda K H
0.318 0.135 0.434
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1534984332
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40