bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0612_orf2
Length=220
Score E
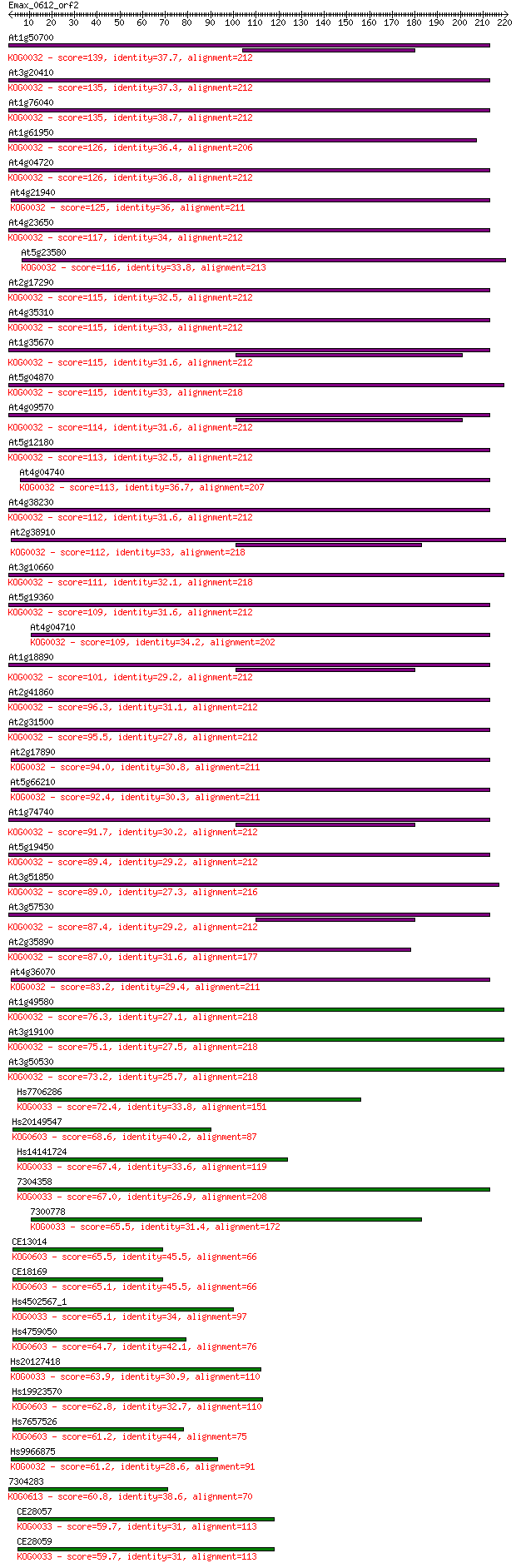
Sequences producing significant alignments: (Bits) Value
At1g50700 139 6e-33
At3g20410 135 6e-32
At1g76040 135 7e-32
At1g61950 126 4e-29
At4g04720 126 4e-29
At4g21940 125 1e-28
At4g23650 117 2e-26
At5g23580 116 4e-26
At2g17290 115 5e-26
At4g35310 115 6e-26
At1g35670 115 6e-26
At5g04870 115 8e-26
At4g09570 114 2e-25
At5g12180 113 3e-25
At4g04740 113 4e-25
At4g38230 112 4e-25
At2g38910 112 8e-25
At3g10660 111 1e-24
At5g19360 109 3e-24
At4g04710 109 4e-24
At1g18890 101 1e-21
At2g41860 96.3 5e-20
At2g31500 95.5 8e-20
At2g17890 94.0 2e-19
At5g66210 92.4 6e-19
At1g74740 91.7 1e-18
At5g19450 89.4 5e-18
At3g51850 89.0 7e-18
At3g57530 87.4 2e-17
At2g35890 87.0 2e-17
At4g36070 83.2 4e-16
At1g49580 76.3 4e-14
At3g19100 75.1 1e-13
At3g50530 73.2 4e-13
Hs7706286 72.4 6e-13
Hs20149547 68.6 9e-12
Hs14141724 67.4 2e-11
7304358 67.0 3e-11
7300778 65.5 8e-11
CE13014 65.5 9e-11
CE18169 65.1 1e-10
Hs4502567_1 65.1 1e-10
Hs4759050 64.7 1e-10
Hs20127418 63.9 2e-10
Hs19923570 62.8 5e-10
Hs7657526 61.2 1e-09
Hs9966875 61.2 2e-09
7304283 60.8 2e-09
CE28057 59.7 4e-09
CE28059 59.7 5e-09
> At1g50700
Length=521
Score = 139 bits (349), Expect = 6e-33, Method: Compositional matrix adjust.
Identities = 80/212 (37%), Positives = 113/212 (53%), Gaps = 15/212 (7%)
Query 1 GQTDQEILKRVERGKFSFEMPDWGHVSEEAKDLVKKMLTYDPNKRINAEECLHHPWMTKF 60
+T++ I + G+ FE W +S AKDLV++MLT DP +RI+A E L HPW+ +
Sbjct 275 AETEKGIFDAILEGEIDFESQPWPSISNSAKDLVRRMLTQDPKRRISAAEVLKHPWLREG 334
Query 61 ASHKDTSINKHVLTGALGNMKKFQATQKLAQAAILFMGSKLTTVEETKELTTIFRQLDKN 120
D I+ VL+ MK+F+A KL + A+ + + T EE + L +F +D +
Sbjct 335 GEASDKPIDSAVLS----RMKQFRAMNKLKKLALKVIAENIDT-EEIQGLKAMFANIDTD 389
Query 121 GDGQLDRKELIEGYRKLLDWKGESGEVDEATIEQEVDQILATVDFDKNGYIEYSEFVTVC 180
G + +EL EG KL E EV Q++ D D NG I+Y EF+T
Sbjct 390 NSGTITYEELKEGLAKL----------GSRLTEAEVKQLMDAADVDGNGSIDYIEFITAT 439
Query 181 MDKQLLLSRERLLQAFQQFDSDGSGKITNDEL 212
M + L S E + +AFQ FD DGSG IT DEL
Sbjct 440 MHRHRLESNENVYKAFQHFDKDGSGYITTDEL 471
Score = 35.0 bits (79), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 24/76 (31%), Positives = 37/76 (48%), Gaps = 11/76 (14%)
Query 104 VEETKELTTIFRQLDKNGDGQLDRKELIEGYRKLLDWKGESGEVDEATIEQEVDQILATV 163
+E + + F+ DK+G G + EL + E G D+ATI++ IL+ V
Sbjct 445 LESNENVYKAFQHFDKDGSGYITTDELEAALK-------EYGMGDDATIKE----ILSDV 493
Query 164 DFDKNGYIEYSEFVTV 179
D D +G I Y EF +
Sbjct 494 DADNDGRINYDEFCAM 509
> At3g20410
Length=541
Score = 135 bits (340), Expect = 6e-32, Method: Compositional matrix adjust.
Identities = 79/212 (37%), Positives = 111/212 (52%), Gaps = 15/212 (7%)
Query 1 GQTDQEILKRVERGKFSFEMPDWGHVSEEAKDLVKKMLTYDPNKRINAEECLHHPWMTKF 60
+T++ I + G FE W +S AKDLV++MLT DP +RI+A + L HPW+ +
Sbjct 293 AETEKGIFDAILEGHIDFESQPWPSISSSAKDLVRRMLTADPKRRISAADVLQHPWLREG 352
Query 61 ASHKDTSINKHVLTGALGNMKKFQATQKLAQAAILFMGSKLTTVEETKELTTIFRQLDKN 120
D I+ VL+ MK+F+A KL + A+ + + T EE + L +F +D +
Sbjct 353 GEASDKPIDSAVLS----RMKQFRAMNKLKKLALKVIAENIDT-EEIQGLKAMFANIDTD 407
Query 121 GDGQLDRKELIEGYRKLLDWKGESGEVDEATIEQEVDQILATVDFDKNGYIEYSEFVTVC 180
G + +EL EG KL E EV Q++ D D NG I+Y EF+T
Sbjct 408 NSGTITYEELKEGLAKL----------GSKLTEAEVKQLMDAADVDGNGSIDYIEFITAT 457
Query 181 MDKQLLLSRERLLQAFQQFDSDGSGKITNDEL 212
M + L S E L +AFQ FD D SG IT DEL
Sbjct 458 MHRHRLESNENLYKAFQHFDKDSSGYITIDEL 489
> At1g76040
Length=534
Score = 135 bits (340), Expect = 7e-32, Method: Compositional matrix adjust.
Identities = 82/213 (38%), Positives = 113/213 (53%), Gaps = 18/213 (8%)
Query 1 GQTDQEILKRVERGKFSFEMPDWGHVSEEAKDLVKKMLTYDPNKRINAEECLHHPWMTKF 60
G+T++ I + + GK E W +SE AKDL++KML DP KRI A E L HPWMT
Sbjct 287 GETEKTIFEAILEGKLDLETSPWPTISESAKDLIRKMLIRDPKKRITAAEALEHPWMT-- 344
Query 61 ASHKDTSI-NKHVLTGALGNMKKFQATQKLAQAAILFMGSKLTTVEETKELTTIFRQLDK 119
DT I +K + + L MK+F+A KL + A+ + L+ EE K L F+ +D
Sbjct 345 ----DTKISDKPINSAVLVRMKQFRAMNKLKKLALKVIAENLSE-EEIKGLKQTFKNMDT 399
Query 120 NGDGQLDRKELIEGYRKLLDWKGESGEVDEATIEQEVDQILATVDFDKNGYIEYSEFVTV 179
+ G + EL G +L E E+ Q++ D DK+G I+Y EFVT
Sbjct 400 DESGTITFDELRNGLHRL----------GSKLTESEIKQLMEAADVDKSGTIDYIEFVTA 449
Query 180 CMDKQLLLSRERLLQAFQQFDSDGSGKITNDEL 212
M + L E L++AF+ FD D SG IT DEL
Sbjct 450 TMHRHRLEKEENLIEAFKYFDKDRSGFITRDEL 482
> At1g61950
Length=547
Score = 126 bits (316), Expect = 4e-29, Method: Compositional matrix adjust.
Identities = 75/206 (36%), Positives = 105/206 (50%), Gaps = 15/206 (7%)
Query 1 GQTDQEILKRVERGKFSFEMPDWGHVSEEAKDLVKKMLTYDPNKRINAEECLHHPWMTKF 60
+TD+ I + + RG+ FE W +SE AKDLV+ ML YDP KR A + L HPW+ +
Sbjct 301 AETDKGIFEEILRGEIDFESEPWPSISESAKDLVRNMLKYDPKKRFTAAQVLEHPWIREG 360
Query 61 ASHKDTSINKHVLTGALGNMKKFQATQKLAQAAILFMGSKLTTVEETKELTTIFRQLDKN 120
D I+ VL+ MK+ +A KL + A F+ L EE K L T+F +D +
Sbjct 361 GEASDKPIDSAVLS----RMKQLRAMNKLKKLAFKFIAQNLKE-EELKGLKTMFANMDTD 415
Query 121 GDGQLDRKELIEGYRKLLDWKGESGEVDEATIEQEVDQILATVDFDKNGYIEYSEFVTVC 180
G + EL G KL E EV Q+L D D NG I+Y EF++
Sbjct 416 KSGTITYDELKSGLEKL----------GSRLTETEVKQLLEDADVDGNGTIDYIEFISAT 465
Query 181 MDKQLLLSRERLLQAFQQFDSDGSGK 206
M++ + + L +AFQ FD D SG+
Sbjct 466 MNRFRVEREDNLFKAFQHFDKDNSGQ 491
> At4g04720
Length=531
Score = 126 bits (316), Expect = 4e-29, Method: Compositional matrix adjust.
Identities = 78/212 (36%), Positives = 111/212 (52%), Gaps = 16/212 (7%)
Query 1 GQTDQEILKRVERGKFSFEMPDWGHVSEEAKDLVKKMLTYDPNKRINAEECLHHPWMTKF 60
+ ++ I V +G+ F W +SE AKDLV+KMLT DP +RI A + L HPW+ K
Sbjct 282 AENEKGIFDEVIKGEIDFVSEPWPSISESAKDLVRKMLTKDPKRRITAAQVLEHPWI-KG 340
Query 61 ASHKDTSINKHVLTGALGNMKKFQATQKLAQAAILFMGSKLTTVEETKELTTIFRQLDKN 120
D I+ VL+ MK+F+A KL + A+ + L+ EE K L T+F +D +
Sbjct 341 GEAPDKPIDSAVLS----RMKQFRAMNKLKKLALKVIAESLSE-EEIKGLKTMFANIDTD 395
Query 121 GDGQLDRKELIEGYRKLLDWKGESGEVDEATIEQEVDQILATVDFDKNGYIEYSEFVTVC 180
G + +EL G +L E+ EV Q++ D D NG I+Y EF++
Sbjct 396 KSGTITYEELKTGLTRLGSRLSET----------EVKQLMEAADVDGNGTIDYYEFISAT 445
Query 181 MDKQLLLSRERLLQAFQQFDSDGSGKITNDEL 212
M + L E + +AFQ FD D SG IT DEL
Sbjct 446 MHRYKLDRDEHVYKAFQHFDKDNSGHITRDEL 477
> At4g21940
Length=554
Score = 125 bits (313), Expect = 1e-28, Method: Compositional matrix adjust.
Identities = 76/211 (36%), Positives = 112/211 (53%), Gaps = 16/211 (7%)
Query 2 QTDQEILKRVERGKFSFEMPDWGHVSEEAKDLVKKMLTYDPNKRINAEECLHHPWMTKFA 61
+T++ I + +G+ F+ W +SE AKDLV+K+LT DP +RI+A + L HPW+ +
Sbjct 305 ETEKGIFNEIIKGEIDFDSQPWPSISESAKDLVRKLLTKDPKQRISAAQALEHPWI-RGG 363
Query 62 SHKDTSINKHVLTGALGNMKKFQATQKLAQAAILFMGSKLTTVEETKELTTIFRQLDKNG 121
D I+ VL+ MK+F+A KL + A+ + L+ EE K L T+F +D +
Sbjct 364 EAPDKPIDSAVLS----RMKQFRAMNKLKKLALKVIAESLSE-EEIKGLKTMFANMDTDK 418
Query 122 DGQLDRKELIEGYRKLLDWKGESGEVDEATIEQEVDQILATVDFDKNGYIEYSEFVTVCM 181
G + +EL G KL E EV Q++ D D NG I+Y EF++ M
Sbjct 419 SGTITYEELKNGLAKL----------GSKLTEAEVKQLMEAADVDGNGTIDYIEFISATM 468
Query 182 DKQLLLSRERLLQAFQQFDSDGSGKITNDEL 212
+ E + +AFQ FD D SG IT DEL
Sbjct 469 HRYRFDRDEHVFKAFQYFDKDNSGFITMDEL 499
> At4g23650
Length=529
Score = 117 bits (292), Expect = 2e-26, Method: Compositional matrix adjust.
Identities = 72/212 (33%), Positives = 112/212 (52%), Gaps = 15/212 (7%)
Query 1 GQTDQEILKRVERGKFSFEMPDWGHVSEEAKDLVKKMLTYDPNKRINAEECLHHPWMTKF 60
G+ + I + +G+ F W +S+ AKDLV+KML YDP R+ A E L+HPW+ +
Sbjct 280 GENETGIFDAILQGQLDFSADPWPALSDGAKDLVRKMLKYDPKDRLTAAEVLNHPWIRED 339
Query 61 ASHKDTSINKHVLTGALGNMKKFQATQKLAQAAILFMGSKLTTVEETKELTTIFRQLDKN 120
D ++ VL+ MK+F+A KL + A+ + L+ EE L +F+ LD +
Sbjct 340 GEASDKPLDNAVLS----RMKQFRAMNKLKKMALKVIAENLSE-EEIIGLKEMFKSLDTD 394
Query 121 GDGQLDRKELIEGYRKLLDWKGESGEVDEATIEQEVDQILATVDFDKNGYIEYSEFVTVC 180
+G + +EL G KL ++ EA E+ Q++ D D +G I+Y EF++
Sbjct 395 NNGIVTLEELRTGLPKL------GSKISEA----EIRQLMEAADMDGDGSIDYLEFISAT 444
Query 181 MDKQLLLSRERLLQAFQQFDSDGSGKITNDEL 212
M + + L AFQ FD+D SG IT +EL
Sbjct 445 MHMNRIEREDHLYTAFQFFDNDNSGYITMEEL 476
> At5g23580
Length=490
Score = 116 bits (290), Expect = 4e-26, Method: Compositional matrix adjust.
Identities = 72/216 (33%), Positives = 112/216 (51%), Gaps = 18/216 (8%)
Query 7 ILKRVERGKFSFEMPDWGHVSEEAKDLVKKMLTYDPNKRINAEECLHHPWMTKFASHKDT 66
I +++ +GK FE+ W +SE AKDL+KKML +P KR+ A + L HPW+ D
Sbjct 230 IFRKILQGKLEFEINPWPSISESAKDLIKKMLESNPKKRLTAHQVLCHPWIVDDKVAPDK 289
Query 67 SINKHVLTGALGNMKKFQATQKLAQAAILFMGSKLTTVEETKELTTIFRQLDKNGDGQLD 126
++ V++ +KKF A KL + A+ + +L+ EE L +F+ +D + G +
Sbjct 290 PLDCAVVS----RLKKFSAMNKLKKMALRVIAERLSE-EEIGGLKELFKMIDTDKSGTIT 344
Query 127 RKELIEGYRKLLDWKGESGEVDEATIEQEVDQILATVDFDKNGYIEYSEFVTVCMDKQLL 186
+EL + R+ V +E E+ ++L D D++G I+Y EF+ + L
Sbjct 345 FEELKDSMRR----------VGSELMESEIQELLRAADVDESGTIDYGEFLAATIHLNKL 394
Query 187 LSRERLLQAFQQFDSDGSGKITNDELA---KLFGIT 219
E L+ AF FD D SG IT +EL K FGI
Sbjct 395 EREENLVAAFSFFDKDASGYITIEELQQAWKEFGIN 430
> At2g17290
Length=544
Score = 115 bits (289), Expect = 5e-26, Method: Compositional matrix adjust.
Identities = 69/212 (32%), Positives = 105/212 (49%), Gaps = 15/212 (7%)
Query 1 GQTDQEILKRVERGKFSFEMPDWGHVSEEAKDLVKKMLTYDPNKRINAEECLHHPWMTKF 60
+T Q I V +G F+ W +S+ AKDL++KML P++R+ A E L HPW+ +
Sbjct 287 AETQQGIFDAVLKGYIDFDTDPWPVISDSAKDLIRKMLCSSPSERLTAHEVLRHPWICEN 346
Query 61 ASHKDTSINKHVLTGALGNMKKFQATQKLAQAAILFMGSKLTTVEETKELTTIFRQLDKN 120
D +++ VL+ +K+F A KL + A+ + L+ EE L +F +D +
Sbjct 347 GVAPDRALDPAVLS----RLKQFSAMNKLKKMALKVIAESLSE-EEIAGLRAMFEAMDTD 401
Query 121 GDGQLDRKELIEGYRKLLDWKGESGEVDEATIEQEVDQILATVDFDKNGYIEYSEFVTVC 180
G + EL G R+ + E+ ++ D D +G I+YSEF+
Sbjct 402 NSGAITFDELKAGLRRY----------GSTLKDTEIRDLMEAADVDNSGTIDYSEFIAAT 451
Query 181 MDKQLLLSRERLLQAFQQFDSDGSGKITNDEL 212
+ L E L+ AFQ FD DGSG IT DEL
Sbjct 452 IHLNKLEREEHLVSAFQYFDKDGSGYITIDEL 483
> At4g35310
Length=556
Score = 115 bits (288), Expect = 6e-26, Method: Compositional matrix adjust.
Identities = 70/212 (33%), Positives = 106/212 (50%), Gaps = 15/212 (7%)
Query 1 GQTDQEILKRVERGKFSFEMPDWGHVSEEAKDLVKKMLTYDPNKRINAEECLHHPWMTKF 60
+T Q I V +G FE W +S+ AKDL+++ML+ P +R+ A E L HPW+ +
Sbjct 299 AETQQGIFDAVLKGYIDFESDPWPVISDSAKDLIRRMLSSKPAERLTAHEVLRHPWICEN 358
Query 61 ASHKDTSINKHVLTGALGNMKKFQATQKLAQAAILFMGSKLTTVEETKELTTIFRQLDKN 120
D +++ VL+ +K+F A KL + A+ + L+ EE L +F+ +D +
Sbjct 359 GVAPDRALDPAVLS----RLKQFSAMNKLKKMALKVIAESLSE-EEIAGLREMFQAMDTD 413
Query 121 GDGQLDRKELIEGYRKLLDWKGESGEVDEATIEQEVDQILATVDFDKNGYIEYSEFVTVC 180
G + EL G RK + E+ ++ D D +G I+YSEF+
Sbjct 414 NSGAITFDELKAGLRKY----------GSTLKDTEIHDLMDAADVDNSGTIDYSEFIAAT 463
Query 181 MDKQLLLSRERLLQAFQQFDSDGSGKITNDEL 212
+ L E L+ AFQ FD DGSG IT DEL
Sbjct 464 IHLNKLEREEHLVAAFQYFDKDGSGFITIDEL 495
> At1g35670
Length=495
Score = 115 bits (288), Expect = 6e-26, Method: Compositional matrix adjust.
Identities = 67/212 (31%), Positives = 108/212 (50%), Gaps = 15/212 (7%)
Query 1 GQTDQEILKRVERGKFSFEMPDWGHVSEEAKDLVKKMLTYDPNKRINAEECLHHPWMTKF 60
+T+ I +++ +GK F+ W +SE AKDL+ KML P KRI+A E L HPW+
Sbjct 228 AETESGIFRQILQGKLDFKSDPWPTISEAAKDLIYKMLERSPKKRISAHEALCHPWIVDE 287
Query 61 ASHKDTSINKHVLTGALGNMKKFQATQKLAQAAILFMGSKLTTVEETKELTTIFRQLDKN 120
+ D ++ VL+ +K+F K+ + A+ + +L+ EE L +F+ +D +
Sbjct 288 QAAPDKPLDPAVLS----RLKQFSQMNKIKKMALRVIAERLSE-EEIGGLKELFKMIDTD 342
Query 121 GDGQLDRKELIEGYRKLLDWKGESGEVDEATIEQEVDQILATVDFDKNGYIEYSEFVTVC 180
G + +EL G ++ V +E E+ ++ D D +G I+Y EF+
Sbjct 343 NSGTITFEELKAGLKR----------VGSELMESEIKSLMDAADIDNSGTIDYGEFLAAT 392
Query 181 MDKQLLLSRERLLQAFQQFDSDGSGKITNDEL 212
+ + E L+ AF FD DGSG IT DEL
Sbjct 393 LHMNKMEREENLVAAFSYFDKDGSGYITIDEL 424
Score = 31.6 bits (70), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 24/100 (24%), Positives = 42/100 (42%), Gaps = 13/100 (13%)
Query 101 LTTVEETKELTTIFRQLDKNGDGQLDRKELIEGYRKLLDWKGESGEVDEATIEQEVDQIL 160
+ +E + L F DK+G G + EL +S + + +D ++
Sbjct 395 MNKMEREENLVAAFSYFDKDGSGYITIDEL------------QSACTEFGLCDTPLDDMI 442
Query 161 ATVDFDKNGYIEYSEFVTVCMDKQLLLSRERLLQAFQQFD 200
+D D +G I++SEF T M K + R R + F+
Sbjct 443 KEIDLDNDGKIDFSEF-TAMMRKGDGVGRSRTMMKNLNFN 481
> At5g04870
Length=610
Score = 115 bits (288), Expect = 8e-26, Method: Compositional matrix adjust.
Identities = 72/221 (32%), Positives = 107/221 (48%), Gaps = 18/221 (8%)
Query 1 GQTDQEILKRVERGKFSFEMPDWGHVSEEAKDLVKKMLTYDPNKRINAEECLHHPWMTKF 60
+T+Q I ++V G F W +SE AKDLV+KML DP KR+ A + L HPW+
Sbjct 352 AETEQGIFEQVLHGDLDFSSDPWPSISESAKDLVRKMLVRDPKKRLTAHQVLCHPWVQVD 411
Query 61 ASHKDTSINKHVLTGALGNMKKFQATQKLAQAAILFMGSKLTTVEETKELTTIFRQLDKN 120
D ++ VL+ MK+F A K + A+ + L+ EE L +F +D +
Sbjct 412 GVAPDKPLDSAVLS----RMKQFSAMNKFKKMALRVIAESLSE-EEIAGLKEMFNMIDAD 466
Query 121 GDGQLDRKELIEGYRKLLDWKGESGEVDEATIEQEVDQILATVDFDKNGYIEYSEFVTVC 180
GQ+ +EL G ++ V E E+ ++ D D +G I+Y EF+
Sbjct 467 KSGQITFEELKAGLKR----------VGANLKESEILDLMQAADVDNSGTIDYKEFIAAT 516
Query 181 MDKQLLLSRERLLQAFQQFDSDGSGKITNDELAKL---FGI 218
+ + + L AF FD DGSG IT DEL + FG+
Sbjct 517 LHLNKIEREDHLFAAFTYFDKDGSGYITPDELQQACEEFGV 557
> At4g09570
Length=501
Score = 114 bits (284), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 67/212 (31%), Positives = 108/212 (50%), Gaps = 15/212 (7%)
Query 1 GQTDQEILKRVERGKFSFEMPDWGHVSEEAKDLVKKMLTYDPNKRINAEECLHHPWMTKF 60
+T+ I +++ +GK F+ W +SE AKDL+ KML P KRI+A E L HPW+
Sbjct 227 AETESGIFRQILQGKIDFKSDPWPTISEGAKDLIYKMLDRSPKKRISAHEALCHPWIVDE 286
Query 61 ASHKDTSINKHVLTGALGNMKKFQATQKLAQAAILFMGSKLTTVEETKELTTIFRQLDKN 120
+ D ++ VL+ +K+F K+ + A+ + +L+ EE L +F+ +D +
Sbjct 287 HAAPDKPLDPAVLS----RLKQFSQMNKIKKMALRVIAERLSE-EEIGGLKELFKMIDTD 341
Query 121 GDGQLDRKELIEGYRKLLDWKGESGEVDEATIEQEVDQILATVDFDKNGYIEYSEFVTVC 180
G + +EL G ++ V +E E+ ++ D D +G I+Y EF+
Sbjct 342 NSGTITFEELKAGLKR----------VGSELMESEIKSLMDAADIDNSGTIDYGEFLAAT 391
Query 181 MDKQLLLSRERLLQAFQQFDSDGSGKITNDEL 212
+ + E L+ AF FD DGSG IT DEL
Sbjct 392 LHINKMEREENLVVAFSYFDKDGSGYITIDEL 423
Score = 30.8 bits (68), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 23/100 (23%), Positives = 42/100 (42%), Gaps = 13/100 (13%)
Query 101 LTTVEETKELTTIFRQLDKNGDGQLDRKELIEGYRKLLDWKGESGEVDEATIEQEVDQIL 160
+ +E + L F DK+G G + EL + + + +D ++
Sbjct 394 INKMEREENLVVAFSYFDKDGSGYITIDELQQACTEF------------GLCDTPLDDMI 441
Query 161 ATVDFDKNGYIEYSEFVTVCMDKQLLLSRERLLQAFQQFD 200
+D D +G I++SEF T M K + R R ++ F+
Sbjct 442 KEIDLDNDGKIDFSEF-TAMMKKGDGVGRSRTMRNNLNFN 480
> At5g12180
Length=528
Score = 113 bits (282), Expect = 3e-25, Method: Compositional matrix adjust.
Identities = 69/212 (32%), Positives = 105/212 (49%), Gaps = 15/212 (7%)
Query 1 GQTDQEILKRVERGKFSFEMPDWGHVSEEAKDLVKKMLTYDPNKRINAEECLHHPWMTKF 60
+++ I + RG F W +S +AKDLVKKML DP +R+ A + L+HPW+ +
Sbjct 275 AESENGIFNAILRGHVDFSSDPWPSISPQAKDLVKKMLNSDPKQRLTAAQVLNHPWIKED 334
Query 61 ASHKDTSINKHVLTGALGNMKKFQATQKLAQAAILFMGSKLTTVEETKELTTIFRQLDKN 120
D ++ V++ +K+F+A + A+ + L+ EE L +F+ +D +
Sbjct 335 GEAPDVPLDNAVMS----RLKQFKAMNNFKKVALRVIAGCLSE-EEIMGLKEMFKGMDTD 389
Query 121 GDGQLDRKELIEGYRKLLDWKGESGEVDEATIEQEVDQILATVDFDKNGYIEYSEFVTVC 180
G + +EL +G K +G E EV Q++ D D NG I+Y EF+
Sbjct 390 SSGTITLEELRQGLAK----QGTR------LSEYEVQQLMEAADADGNGTIDYGEFIAAT 439
Query 181 MDKQLLLSRERLLQAFQQFDSDGSGKITNDEL 212
M L E L AFQ FD D SG IT +EL
Sbjct 440 MHINRLDREEHLYSAFQHFDKDNSGYITMEEL 471
> At4g04740
Length=520
Score = 113 bits (282), Expect = 4e-25, Method: Compositional matrix adjust.
Identities = 76/207 (36%), Positives = 106/207 (51%), Gaps = 20/207 (9%)
Query 6 EILKRVERGKFSFEMPDWGHVSEEAKDLVKKMLTYDPNKRINAEECLHHPWMTKFASHKD 65
EILK K F W +S+ AKDLV+KMLT DP +RI A + L HPW+ K +
Sbjct 280 EILK----CKIDFVREPWPSISDSAKDLVEKMLTEDPKRRITAAQVLEHPWI-KGGEAPE 334
Query 66 TSINKHVLTGALGNMKKFQATQKLAQAAILFMGSKLTTVEETKELTTIFRQLDKNGDGQL 125
I+ VL+ MK+F+A KL + A+ L+ EE K L T+F +D N G +
Sbjct 335 KPIDSTVLS----RMKQFRAMNKLKKLALKVSAVSLSE-EEIKGLKTLFANMDTNRSGTI 389
Query 126 DRKELIEGYRKLLDWKGESGEVDEATIEQEVDQILATVDFDKNGYIEYSEFVTVCMDKQL 185
++L G +L E+ EV Q++ D D NG I+Y EF++ M +
Sbjct 390 TYEQLQTGLSRLRSRLSET----------EVQQLVEASDVDGNGTIDYYEFISATMHRYK 439
Query 186 LLSRERLLQAFQQFDSDGSGKITNDEL 212
L E + +AFQ D D +G IT DEL
Sbjct 440 LHHDEHVHKAFQHLDKDKNGHITRDEL 466
> At4g38230
Length=484
Score = 112 bits (281), Expect = 4e-25, Method: Compositional matrix adjust.
Identities = 67/212 (31%), Positives = 105/212 (49%), Gaps = 15/212 (7%)
Query 1 GQTDQEILKRVERGKFSFEMPDWGHVSEEAKDLVKKMLTYDPNKRINAEECLHHPWMTKF 60
+T Q I V +G F+ W +S+ AK+L++ ML P++R+ A + L HPW+ +
Sbjct 226 AETQQGIFDAVLKGHIDFDSDPWPLISDSAKNLIRGMLCSRPSERLTAHQVLRHPWICEN 285
Query 61 ASHKDTSINKHVLTGALGNMKKFQATQKLAQAAILFMGSKLTTVEETKELTTIFRQLDKN 120
D +++ VL+ +K+F A KL Q A+ + L+ EE L +F+ +D +
Sbjct 286 GVAPDRALDPAVLS----RLKQFSAMNKLKQMALRVIAESLSE-EEIAGLKEMFKAMDTD 340
Query 121 GDGQLDRKELIEGYRKLLDWKGESGEVDEATIEQEVDQILATVDFDKNGYIEYSEFVTVC 180
G + EL G R+ + E+ ++ D DK+G I+Y EF+
Sbjct 341 NSGAITFDELKAGLRRY----------GSTLKDTEIRDLMEAADIDKSGTIDYGEFIAAT 390
Query 181 MDKQLLLSRERLLQAFQQFDSDGSGKITNDEL 212
+ L E LL AF+ FD DGSG IT DEL
Sbjct 391 IHLNKLEREEHLLSAFRYFDKDGSGYITIDEL 422
> At2g38910
Length=583
Score = 112 bits (279), Expect = 8e-25, Method: Compositional matrix adjust.
Identities = 72/222 (32%), Positives = 111/222 (50%), Gaps = 20/222 (9%)
Query 2 QTDQEILKRVERGKFSFEMPDWGHVSEEAKDLVKKMLTYDPNKRINAEECLHHPWMTKFA 61
+T+Q I ++V +G F W VSE AKDLV++ML DP KR+ E L HPW A
Sbjct 337 ETEQGIFEQVLKGDLDFISEPWPSVSESAKDLVRRMLIRDPKKRMTTHEVLCHPW----A 392
Query 62 SHKDTSINKHVLTGALGNMKKFQATQKLAQAAILFMGSKLTTVEETKELTTIFRQLDKNG 121
+++K + + L +++F A KL + AI + L+ EE L +F+ +D +
Sbjct 393 RVDGVALDKPLDSAVLSRLQQFSAMNKLKKIAIKVIAESLSE-EEIAGLKEMFKMIDTDN 451
Query 122 DGQLDRKELIEGYRKL-LDWKGESGEVDEATIEQEVDQILATVDFDKNGYIEYSEFVTVC 180
G + +EL +G ++ D K + E+ ++ D D +G I+Y EF+
Sbjct 452 SGHITLEELKKGLDRVGADLK-----------DSEILGLMQAADIDNSGTIDYGEFIAAM 500
Query 181 MDKQLLLSRERLLQAFQQFDSDGSGKITNDEL---AKLFGIT 219
+ + + L AF FD DGSG IT DEL K FG+
Sbjct 501 VHLNKIEKEDHLFTAFSYFDQDGSGYITRDELQQACKQFGLA 542
Score = 39.3 bits (90), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 25/82 (30%), Positives = 38/82 (46%), Gaps = 12/82 (14%)
Query 101 LTTVEETKELTTIFRQLDKNGDGQLDRKELIEGYRKLLDWKGESGEVDEATIEQEVDQIL 160
L +E+ L T F D++G G + R EL + ++ + +D IL
Sbjct 503 LNKIEKEDHLFTAFSYFDQDGSGYITRDELQQACKQF------------GLADVHLDDIL 550
Query 161 ATVDFDKNGYIEYSEFVTVCMD 182
VD D +G I+YSEFV + D
Sbjct 551 REVDKDNDGRIDYSEFVDMMQD 572
> At3g10660
Length=646
Score = 111 bits (278), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 70/221 (31%), Positives = 107/221 (48%), Gaps = 18/221 (8%)
Query 1 GQTDQEILKRVERGKFSFEMPDWGHVSEEAKDLVKKMLTYDPNKRINAEECLHHPWMTKF 60
+T+Q I ++V G F W +SE AKDLV+KML DP +R+ A + L HPW+
Sbjct 388 AETEQGIFEQVLHGDLDFSSDPWPSISESAKDLVRKMLVRDPKRRLTAHQVLCHPWVQID 447
Query 61 ASHKDTSINKHVLTGALGNMKKFQATQKLAQAAILFMGSKLTTVEETKELTTIFRQLDKN 120
D ++ VL+ MK+F A K + A+ + L+ EE L +F+ +D +
Sbjct 448 GVAPDKPLDSAVLS----RMKQFSAMNKFKKMALRVIAESLSE-EEIAGLKQMFKMIDAD 502
Query 121 GDGQLDRKELIEGYRKLLDWKGESGEVDEATIEQEVDQILATVDFDKNGYIEYSEFVTVC 180
GQ+ +EL G ++ V E E+ ++ D D +G I+Y EF+
Sbjct 503 NSGQITFEELKAGLKR----------VGANLKESEILDLMQAADVDNSGTIDYKEFIAAT 552
Query 181 MDKQLLLSRERLLQAFQQFDSDGSGKITNDELAKL---FGI 218
+ + + L AF FD D SG IT DEL + FG+
Sbjct 553 LHLNKIEREDHLFAAFSYFDKDESGFITPDELQQACEEFGV 593
> At5g19360
Length=523
Score = 109 bits (273), Expect = 3e-24, Method: Compositional matrix adjust.
Identities = 67/212 (31%), Positives = 105/212 (49%), Gaps = 15/212 (7%)
Query 1 GQTDQEILKRVERGKFSFEMPDWGHVSEEAKDLVKKMLTYDPNKRINAEECLHHPWMTKF 60
+++ I + G+ F W +S +AKDLV+KML DP +R+ A + L+HPW+ +
Sbjct 270 AESENGIFNAILSGQVDFSSDPWPVISPQAKDLVRKMLNSDPKQRLTAAQVLNHPWIKED 329
Query 61 ASHKDTSINKHVLTGALGNMKKFQATQKLAQAAILFMGSKLTTVEETKELTTIFRQLDKN 120
D ++ V++ +K+F+A + A+ + L+ EE L +F+ +D +
Sbjct 330 GEAPDVPLDNAVMS----RLKQFKAMNNFKKVALRVIAGCLSE-EEIMGLKEMFKGMDTD 384
Query 121 GDGQLDRKELIEGYRKLLDWKGESGEVDEATIEQEVDQILATVDFDKNGYIEYSEFVTVC 180
G + +EL +G K +G E EV Q++ D D NG I+Y EF+
Sbjct 385 NSGTITLEELRQGLAK----QGTR------LSEYEVQQLMEAADADGNGTIDYGEFIAAT 434
Query 181 MDKQLLLSRERLLQAFQQFDSDGSGKITNDEL 212
M L E L AFQ FD D SG IT +EL
Sbjct 435 MHINRLDREEHLYSAFQHFDKDNSGYITTEEL 466
> At4g04710
Length=575
Score = 109 bits (273), Expect = 4e-24, Method: Compositional matrix adjust.
Identities = 69/202 (34%), Positives = 107/202 (52%), Gaps = 15/202 (7%)
Query 11 VERGKFSFEMPDWGHVSEEAKDLVKKMLTYDPNKRINAEECLHHPWMTKFASHKDTSINK 70
++ + FE W +S +AK L+ KMLT P +RI+A + L HPWM K + +K
Sbjct 245 IKECRLDFESQPWPLISFKAKHLIGKMLTKKPKERISAADVLEHPWM------KSEAPDK 298
Query 71 HVLTGALGNMKKFQATQKLAQAAILFMGSKLTTVEETKELTTIFRQLDKNGDGQLDRKEL 130
+ L MK+F+A KL + A+ + L+ EE K L T+F +D + G + +EL
Sbjct 299 PIDNVVLSRMKQFRAMNKLKKLALKVIAEGLSE-EEIKGLKTMFENMDMDKSGSITYEEL 357
Query 131 IEGYRKLLDWKGESGEVDEATIEQEVDQILATVDFDKNGYIEYSEFVTVCMDKQLLLSRE 190
G + ++ E ++Q ++ + A D D NG I+Y EF++ M + L E
Sbjct 358 KMGLNR------HGSKLSETEVKQLMEAVSA--DVDGNGTIDYIEFISATMHRHRLERDE 409
Query 191 RLLQAFQQFDSDGSGKITNDEL 212
L +AFQ FD DGSG IT +E+
Sbjct 410 HLYKAFQYFDKDGSGHITKEEV 431
> At1g18890
Length=545
Score = 101 bits (252), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 62/212 (29%), Positives = 103/212 (48%), Gaps = 15/212 (7%)
Query 1 GQTDQEILKRVERGKFSFEMPDWGHVSEEAKDLVKKMLTYDPNKRINAEECLHHPWMTKF 60
+T+Q + + RG F+ W +SE AK LVK+ML DP KR+ A++ L HPW+
Sbjct 265 AETEQGVALAILRGVLDFKRDPWPQISESAKSLVKQMLDPDPTKRLTAQQVLAHPWIQNA 324
Query 61 ASHKDTSINKHVLTGALGNMKKFQATQKLAQAAILFMGSKLTTVEETKELTTIFRQLDKN 120
+ + V + +K+F + + + + L +++E + + +F +D +
Sbjct 325 KKAPNVPLGDIVRS----RLKQFSMMNRFKKKVLRVIAEHL-SIQEVEVIKNMFSLMDDD 379
Query 121 GDGQLDRKELIEGYRKLLDWKGESGEVDEATIEQEVDQILATVDFDKNGYIEYSEFVTVC 180
DG++ EL G +K+ GE E+ ++ D D NG+++Y EFV V
Sbjct 380 KDGKITYPELKAGLQKVGSQLGEP----------EIKMLMEVADVDGNGFLDYGEFVAVI 429
Query 181 MDKQLLLSRERLLQAFQQFDSDGSGKITNDEL 212
+ Q + + E AF FD DGS I DEL
Sbjct 430 IHLQKIENDELFKLAFMFFDKDGSTYIELDEL 461
Score = 32.0 bits (71), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 27/79 (34%), Positives = 37/79 (46%), Gaps = 10/79 (12%)
Query 101 LTTVEETKELTTIFRQLDKNGDGQLDRKELIEGYRKLLDWKGESGEVDEATIEQEVDQIL 160
L +E + F DK+G ++ EL E L D E GE D + + I+
Sbjct 432 LQKIENDELFKLAFMFFDKDGSTYIELDELREA---LAD---ELGEPDASVLSD----IM 481
Query 161 ATVDFDKNGYIEYSEFVTV 179
VD DK+G I Y EFVT+
Sbjct 482 REVDTDKDGRINYDEFVTM 500
> At2g41860
Length=530
Score = 96.3 bits (238), Expect = 5e-20, Method: Compositional matrix adjust.
Identities = 66/213 (30%), Positives = 109/213 (51%), Gaps = 17/213 (7%)
Query 1 GQTDQEILKRVERGKFSFEMPDWGHVSEEAKDLVKKMLTYDPNKRINAEECLHHPWMTKF 60
+T+ + K + + F+ W VS+ AKDL+KKML DP +R+ A++ L HPW+
Sbjct 256 AETEHGVAKAILKSVIDFKRDPWPKVSDNAKDLIKKMLHPDPRRRLTAQQVLDHPWIQNG 315
Query 61 ASHKDTSINKHVLTGALGNMKKFQATQKLAQAAILFMGSKLTTVEETKELTTIFRQLDKN 120
+ + S+ + V +K+F KL + A+ + L +VEET + F+ +D +
Sbjct 316 KNASNVSLGETV----RARLKQFSVMNKLKKRALRVIAEHL-SVEETSCIKERFQVMDTS 370
Query 121 GDGQLDRKELIEGYRKLLDWKGESGEVDEATIEQEVDQILATV-DFDKNGYIEYSEFVTV 179
G++ EL G +KL + Q+ QIL D DK+GY++ +EFV +
Sbjct 371 NRGKITITELGIGLQKL-----------GIVVPQDDIQILMDAGDVDKDGYLDVNEFVAI 419
Query 180 CMDKQLLLSRERLLQAFQQFDSDGSGKITNDEL 212
+ + L + E L +AF FD + SG I +EL
Sbjct 420 SVHIRKLGNDEHLKKAFTFFDKNKSGYIEIEEL 452
> At2g31500
Length=582
Score = 95.5 bits (236), Expect = 8e-20, Method: Compositional matrix adjust.
Identities = 59/212 (27%), Positives = 103/212 (48%), Gaps = 15/212 (7%)
Query 1 GQTDQEILKRVERGKFSFEMPDWGHVSEEAKDLVKKMLTYDPNKRINAEECLHHPWMTKF 60
+T++ I + RG FE W VS EAK+LVK ML +P R+ +E L HPW+
Sbjct 268 AETEEGIAHAIVRGNIDFERDPWPKVSHEAKELVKNMLDANPYSRLTVQEVLEHPWIRNA 327
Query 61 ASHKDTSINKHVLTGALGNMKKFQATQKLAQAAILFMGSKLTTVEETKELTTIFRQLDKN 120
+ ++ +V T +++F + + + + L EE + +F+ +D +
Sbjct 328 ERAPNVNLGDNVRT----KIQQFLLMNRFKKKVLRIVADNLPN-EEIAAIVQMFQTMDTD 382
Query 121 GDGQLDRKELIEGYRKLLDWKGESGEVDEATIEQEVDQILATVDFDKNGYIEYSEFVTVC 180
+G L +EL +G +K + + + +V ++ D D NG + EFVT+
Sbjct 383 KNGHLTFEELRDGLKK----------IGQVVPDGDVKMLMDAADTDGNGMLSCDEFVTLS 432
Query 181 MDKQLLLSRERLLQAFQQFDSDGSGKITNDEL 212
+ + + E L +AF+ FD +G+G I DEL
Sbjct 433 IHLKRMGCDEHLQEAFKYFDKNGNGFIELDEL 464
> At2g17890
Length=571
Score = 94.0 bits (232), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 65/217 (29%), Positives = 104/217 (47%), Gaps = 20/217 (9%)
Query 2 QTDQEILKRVERGKFSFEMPDWGHVSEEAKDLVKKMLTYDPNKRINAEECLHHPWMTKFA 61
+T+ I K V + K F W +S AKD VKK+L DP R+ A + L HPW+ +
Sbjct 313 KTEDGIFKEVLKNKPDFRRKPWPTISNSAKDFVKKLLVKDPRARLTAAQALSHPWVREGG 372
Query 62 SHKDTSINKHVLTGALGNMKKFQATQKLAQAAILFMGSKLTTVEETKELTTIFRQLDKNG 121
+ I+ VL NM++F +L Q A+ + + L EE +L F +D +
Sbjct 373 DASEIPIDISVLN----NMRQFVKFSRLKQFALRALATTLDE-EELADLRDQFDAIDVDK 427
Query 122 DGQLDRKELIEGYRKLLDWKGESGEVDEATIEQEVDQILATVDFDKNGYIEYSEFVTVCM 181
+G + +E+ + K WK + V E IL +D + +G++++ EFV +
Sbjct 428 NGVISLEEMRQALAKDHPWKLKDARVAE---------ILQAIDSNTDGFVDFGEFVAAAL 478
Query 182 DKQLLLS------RERLLQAFQQFDSDGSGKITNDEL 212
L ++R AF++FD DG G IT +EL
Sbjct 479 HVNQLEEHDSEKWQQRSRAAFEKFDIDGDGFITAEEL 515
> At5g66210
Length=523
Score = 92.4 bits (228), Expect = 6e-19, Method: Compositional matrix adjust.
Identities = 64/217 (29%), Positives = 103/217 (47%), Gaps = 20/217 (9%)
Query 2 QTDQEILKRVERGKFSFEMPDWGHVSEEAKDLVKKMLTYDPNKRINAEECLHHPWMTKFA 61
+T+ I K V R K F W +S+ AKD VKK+L DP R+ A + L H W+ +
Sbjct 267 RTEDGIFKEVLRNKPDFSRKPWATISDSAKDFVKKLLVKDPRARLTAAQALSHAWVREGG 326
Query 62 SHKDTSINKHVLTGALGNMKKFQATQKLAQAAILFMGSKLTTVEETKELTTIFRQLDKNG 121
+ D ++ VL N+++F +L Q A+ + S L E +L F +D +
Sbjct 327 NATDIPVDISVLN----NLRQFVRYSRLKQFALRALASTLDEA-EISDLRDQFDAIDVDK 381
Query 122 DGQLDRKELIEGYRKLLDWKGESGEVDEATIEQEVDQILATVDFDKNGYIEYSEFVTVCM 181
+G + +E+ + K L WK + V E IL +D + +G ++++EFV +
Sbjct 382 NGVISLEEMRQALAKDLPWKLKDSRVAE---------ILEAIDSNTDGLVDFTEFVAAAL 432
Query 182 DKQLLLSRE------RLLQAFQQFDSDGSGKITNDEL 212
L + R AF++FD D G IT +EL
Sbjct 433 HVHQLEEHDSEKWQLRSRAAFEKFDLDKDGYITPEEL 469
> At1g74740
Length=567
Score = 91.7 bits (226), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 64/238 (26%), Positives = 106/238 (44%), Gaps = 41/238 (17%)
Query 1 GQTDQEILKRVERGKFSFEMPDWGHVSEEAKDLVKKMLTYDPNKRINAEECL-------- 52
+T+Q + + RG F+ W +SE AK LVK+ML D KR+ A++ L
Sbjct 261 AETEQGVALAILRGVLDFKRDPWSQISESAKSLVKQMLEPDSTKRLTAQQVLGNNAVLVS 320
Query 53 ------------------HHPWMTKFASHKDTSINKHVLTGALGNMKKFQATQKLAQAAI 94
HPW+ + + V + +K+F +L + A+
Sbjct 321 LMEKMAFKVKTVSVSFFADHPWIQNAKKAPNVPLGDIVRS----RLKQFSMMNRLKKKAL 376
Query 95 LFMGSKLTTVEETKELTTIFRQLDKNGDGQLDRKELIEGYRKLLDWKGESGEVDEATIEQ 154
+ L+ ++E + + +F +D + DG++ EL G RK+ GE
Sbjct 377 RVIAEHLS-IQEVEVIRNMFTLMDDDNDGKISYLELRAGLRKVGSQLGEP---------- 425
Query 155 EVDQILATVDFDKNGYIEYSEFVTVCMDKQLLLSRERLLQAFQQFDSDGSGKITNDEL 212
E+ ++ D + NG ++Y EFV V + Q + + E QAF FD DGSG I ++EL
Sbjct 426 EIKLLMEVADVNGNGCLDYGEFVAVIIHLQKMENDEHFRQAFMFFDKDGSGYIESEEL 483
Score = 35.0 bits (79), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 29/79 (36%), Positives = 40/79 (50%), Gaps = 10/79 (12%)
Query 101 LTTVEETKELTTIFRQLDKNGDGQLDRKELIEGYRKLLDWKGESGEVDEATIEQEVDQIL 160
L +E + F DK+G G ++ +EL E L D E GE D + I +D I+
Sbjct 454 LQKMENDEHFRQAFMFFDKDGSGYIESEELREA---LTD---ELGEPDNSVI---ID-IM 503
Query 161 ATVDFDKNGYIEYSEFVTV 179
VD DK+G I Y EFV +
Sbjct 504 REVDTDKDGKINYDEFVVM 522
> At5g19450
Length=533
Score = 89.4 bits (220), Expect = 5e-18, Method: Compositional matrix adjust.
Identities = 62/212 (29%), Positives = 104/212 (49%), Gaps = 14/212 (6%)
Query 1 GQTDQEILKRVERGKFSFEMPDWGHVSEEAKDLVKKMLTYDPNKRINAEECLHHPWMTKF 60
+T+Q + + + R F+ W VSE AKDLV+KML DP KR++A + L H W+
Sbjct 259 AETEQGVAQAIIRSVIDFKRDPWPRVSETAKDLVRKMLEPDPKKRLSAAQVLEHSWIQNA 318
Query 61 ASHKDTSINKHVLTGALGNMKKFQATQKLAQAAILFMGSKLTTVEETKELTTIFRQLDKN 120
+ S+ + V +K+F KL + A+ + L +VEE + F +D
Sbjct 319 KKAPNVSLGETV----KARLKQFSVMNKLKKRALRVIAEHL-SVEEVAGIKEAFEMMDSK 373
Query 121 GDGQLDRKELIEGYRKLLDWKGESGEVDEATIEQEVDQILATVDFDKNGYIEYSEFVTVC 180
G+++ +EL G KL G+ + + ++ ++ D D +G + Y EFV V
Sbjct 374 KTGKINLEELKFGLHKL----GQ-----QQIPDTDLQILMEAADVDGDGTLNYGEFVAVS 424
Query 181 MDKQLLLSRERLLQAFQQFDSDGSGKITNDEL 212
+ + + + E L +AF FD + S I +EL
Sbjct 425 VHLKKMANDEHLHKAFSFFDQNQSDYIEIEEL 456
> At3g51850
Length=503
Score = 89.0 bits (219), Expect = 7e-18, Method: Compositional matrix adjust.
Identities = 59/227 (25%), Positives = 107/227 (47%), Gaps = 26/227 (11%)
Query 1 GQTDQEILKRVERGKFSFEMPDWGHVSEEAKDLVKKMLTYDPNKRINAEECLHHPWMTKF 60
+++Q + + + RG F+ W ++SE AK+LV++ML DP +R+ A++ L HPW+
Sbjct 256 AESEQGVAQAILRGVIDFKREPWPNISETAKNLVRQMLEPDPKRRLTAKQVLEHPWIQNA 315
Query 61 ASHKDTSINKHVLTGALGNMKKFQATQKLAQAAILFMGSKLTTVEETKELTTIFRQLDKN 120
+ + V + +K+F + + A+ + L+T EE +++ +F ++D +
Sbjct 316 KKAPNVPLGDVVKS----RLKQFSVMNRFKRKALRVIAEFLST-EEVEDIKVMFNKMDTD 370
Query 121 GDGQLDRKELIEGYRKLLDWKGESGEVDEATIEQEVDQILATVDFDKNGYIEYSEFVTVC 180
DG + +EL G R ES EV ++ VD G ++Y EFV V
Sbjct 371 NDGIVSIEELKAGLRDFSTQLAES----------EVQMLIEAVDTKGKGTLDYGEFVAVS 420
Query 181 MDKQLLLSR-----------ERLLQAFQQFDSDGSGKITNDELAKLF 216
+ Q L + FQ+ D+D G+I+ +E A +
Sbjct 421 LHLQKELCDALKEDGGDDCVDVANDIFQEVDTDKDGRISYEEFAAMM 467
> At3g57530
Length=560
Score = 87.4 bits (215), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 62/213 (29%), Positives = 108/213 (50%), Gaps = 17/213 (7%)
Query 1 GQTDQEILKRVERGKFSFEMPDWGHVSEEAKDLVKKMLTYDPNKRINAEECLHHPWMTKF 60
+T+Q + + + R F W VSE AKDL++KML D +R+ A++ L HPW+
Sbjct 265 AETEQGVAQAIIRSVLDFRRDPWPKVSENAKDLIRKMLDPDQKRRLTAQQVLDHPWLQNA 324
Query 61 ASHKDTSINKHVLTGALGNMKKFQATQKLAQAAILFMGSKLTTVEETKELTTIFRQLDKN 120
+ + S+ + V +K+F KL + A+ + L+ EE + F+ +D +
Sbjct 325 KTAPNVSLGETV----RARLKQFTVMNKLKKRALRVIAEHLSD-EEASGIREGFQIMDTS 379
Query 121 GDGQLDRKELIEGYRKLLDWKGESGEVDEATIEQEVDQILATV-DFDKNGYIEYSEFVTV 179
G+++ EL G +KL G + I Q+ QIL D D++GY++ EF+ +
Sbjct 380 QRGKINIDELKIGLQKL----GHA-------IPQDDLQILMDAGDIDRDGYLDCDEFIAI 428
Query 180 CMDKQLLLSRERLLQAFQQFDSDGSGKITNDEL 212
+ + + + E L +AF FD + +G I +EL
Sbjct 429 SVHLRKMGNDEHLKKAFAFFDQNNNGYIEIEEL 461
Score = 37.4 bits (85), Expect = 0.025, Method: Compositional matrix adjust.
Identities = 28/70 (40%), Positives = 36/70 (51%), Gaps = 11/70 (15%)
Query 110 LTTIFRQLDKNGDGQLDRKELIEGYRKLLDWKGESGEVDEATIEQEVDQILATVDFDKNG 169
L F D+N +G ++ +EL E L D G S EV VD I+ VD DK+G
Sbjct 441 LKKAFAFFDQNNNGYIEIEELREA---LSDELGTSEEV--------VDAIIRDVDTDKDG 489
Query 170 YIEYSEFVTV 179
I Y EFVT+
Sbjct 490 RISYEEFVTM 499
> At2g35890
Length=520
Score = 87.0 bits (214), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 56/177 (31%), Positives = 91/177 (51%), Gaps = 14/177 (7%)
Query 1 GQTDQEILKRVERGKFSFEMPDWGHVSEEAKDLVKKMLTYDPNKRINAEECLHHPWMTKF 60
G+T++EI V G+ W VSE AKDL++KML +P +R+ A++ L HPW+
Sbjct 334 GETEEEIFNEVLEGELDLTSDPWPQVSESAKDLIRKMLERNPIQRLTAQQVLCHPWIRDE 393
Query 61 ASHKDTSINKHVLTGALGNMKKFQATQKLAQAAILFMGSKLTTVEETKELTTIFRQLDKN 120
+ DT ++ T L +KKF AT KL + A+ + +L+ EE EL F+ +D
Sbjct 394 GNAPDTPLD----TTVLSRLKKFSATDKLKKMALRVIAERLSE-EEIHELRETFKTIDSG 448
Query 121 GDGQLDRKELIEGYRKLLDWKGESGEVDEATIEQEVDQILATVDFDKNGYIEYSEFV 177
G++ KEL G + + +D + I + QI V + ++Y+EF+
Sbjct 449 KSGRVTYKELKNGLERF------NTNLDNSDI-NSLMQIPTDVHLEDT--VDYNEFI 496
> At4g36070
Length=536
Score = 83.2 bits (204), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 62/217 (28%), Positives = 100/217 (46%), Gaps = 20/217 (9%)
Query 2 QTDQEILKRVERGKFSFEMPDWGHVSEEAKDLVKKMLTYDPNKRINAEECLHHPWMTKFA 61
+T I V R K F W +S AKD VKK+L +P R+ A + L H W+ +
Sbjct 273 KTQDGIFNEVMRKKPDFREVPWPTISNGAKDFVKKLLVKEPRARLTAAQALSHSWVKEGG 332
Query 62 SHKDTSINKHVLTGALGNMKKFQATQKLAQAAILFMGSKLTTVEETKELTTIFRQLDKNG 121
+ I+ VL NM++F +L Q A+ + +K +E +L F +D +
Sbjct 333 EASEVPIDISVLN----NMRQFVKFSRLKQIALRAL-AKTINEDELDDLRDQFDAIDIDK 387
Query 122 DGQLDRKELIEGYRKLLDWKGESGEVDEATIEQEVDQILATVDFDKNGYIEYSEFVTVCM 181
+G + +E+ + K + WK + V E IL D + +G ++++EFV +
Sbjct 388 NGSISLEEMRQALAKDVPWKLKDARVAE---------ILQANDSNTDGLVDFTEFVVAAL 438
Query 182 DKQLLLS------RERLLQAFQQFDSDGSGKITNDEL 212
L ++R AF +FD DG G IT +EL
Sbjct 439 HVNQLEEHDSEKWQQRSRAAFDKFDIDGDGFITPEEL 475
> At1g49580
Length=606
Score = 76.3 bits (186), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 59/222 (26%), Positives = 106/222 (47%), Gaps = 18/222 (8%)
Query 1 GQTDQEILKRVERGKFSFEMPDWGHVSEEAKDLVKKMLTYDPNKRINAEECLHHPWMTKF 60
+T+ I + V + SF+ P W +S +AKD VK++L DP +R++A + L HPW+ +
Sbjct 356 ARTESGIFRAVLKADPSFDEPPWPFLSSDAKDFVKRLLFKDPRRRMSASQALMHPWIRAY 415
Query 61 ASHKDTSINKHVLTGALGNMKKFQATQKLAQAAILFMGSKLTTVEETKELTTIFRQLDKN 120
+ D +I +L MK + + L +AA+ + SK +E L T F L N
Sbjct 416 --NTDMNIPFDILI--FRQMKAYLRSSSLRKAALRAL-SKTLIKDEILYLKTQFSLLAPN 470
Query 121 GDGQLDRKELIEGYRKLLDWKGESGEVDEATIEQEVDQILATVDFDKNGYIEYSEFVTVC 180
DG + ++ R L + EA E + + LA ++ + +++ EF
Sbjct 471 KDGLIT----MDTIRMAL-----ASNATEAMKESRIPEFLALLNGLQYRGMDFEEFCAAA 521
Query 181 MD----KQLLLSRERLLQAFQQFDSDGSGKITNDELAKLFGI 218
++ + L + + A++ FD +G+ I +ELA G+
Sbjct 522 INVHQHESLDCWEQSIRHAYELFDKNGNRAIVIEELASELGV 563
> At3g19100
Length=599
Score = 75.1 bits (183), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 60/222 (27%), Positives = 105/222 (47%), Gaps = 19/222 (8%)
Query 1 GQTDQEILKRVERGKFSFEMPDWGHVSEEAKDLVKKMLTYDPNKRINAEECLHHPWMTKF 60
+T+ I + V + SF+ P W +S EAKD VK++L DP KR+ A + L HPW+
Sbjct 350 ARTESGIFRAVLKADPSFDEPPWPSLSFEAKDFVKRLLYKDPRKRMTASQALMHPWI--- 406
Query 61 ASHKDTSINKHVLTGALGNMKKFQATQKLAQAAILFMGSKLTTVEETKELTTIFRQLDKN 120
A +K I +L +K + + L +AA++ + LTT +E L F L N
Sbjct 407 AGYKKIDIPFDILI--FKQIKAYLRSSSLRKAALMALSKTLTT-DELLYLKAQFAHLAPN 463
Query 121 GDGQLDRKELIEGYRKLLDWKGESGEVDEATIEQEVDQILATVDFDKNGYIEYSEF---- 176
+G + ++ R L + EA E + LA ++ + +++ EF
Sbjct 464 KNGLIT----LDSIRLAL-----ATNATEAMKESRIPDFLALLNGLQYKGMDFEEFCAAS 514
Query 177 VTVCMDKQLLLSRERLLQAFQQFDSDGSGKITNDELAKLFGI 218
++V + L + + A++ F+ +G+ I +ELA G+
Sbjct 515 ISVHQHESLDCWEQSIRHAYELFEMNGNRVIVIEELASELGV 556
> At3g50530
Length=601
Score = 73.2 bits (178), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 56/222 (25%), Positives = 102/222 (45%), Gaps = 19/222 (8%)
Query 1 GQTDQEILKRVERGKFSFEMPDWGHVSEEAKDLVKKMLTYDPNKRINAEECLHHPWMTKF 60
+T+ I + V + SF+ P W +S EA+D VK++L DP KR+ A + L HPW+
Sbjct 354 ARTESGIFRAVLKADPSFDDPPWPLLSSEARDFVKRLLNKDPRKRLTAAQALSHPWIK-- 411
Query 61 ASHKDTSINKHVLTGALGNMKKFQATQKLAQAAILFMGSKLTTVEETKELTTIFRQLDKN 120
D + +L L M+ + + L +AA+ + SK TV+E L F L+ +
Sbjct 412 -DSNDAKVPMDILVFKL--MRAYLRSSSLRKAALRAL-SKTLTVDELFYLREQFALLEPS 467
Query 121 GDGQLDRKELIEGYRKLLDWKGESGEVDEATIEQEVDQILATVDFDKNGYIEYSEFVTVC 180
+G + + + K+ +A + + + L + + +++ EF
Sbjct 468 KNGTISLENIKSALMKM---------ATDAMKDSRIPEFLGQLSALQYRRMDFEEFCAAA 518
Query 181 MDKQLLLSRERLLQ----AFQQFDSDGSGKITNDELAKLFGI 218
+ L + +R Q A++ F+ +G+ I DELA G+
Sbjct 519 LSVHQLEALDRWEQHARCAYELFEKEGNRPIMIDELASELGL 560
> Hs7706286
Length=489
Score = 72.4 bits (176), Expect = 6e-13, Method: Compositional matrix adjust.
Identities = 51/151 (33%), Positives = 75/151 (49%), Gaps = 17/151 (11%)
Query 5 QEILKRVERGKFSFEMPDWGHVSEEAKDLVKKMLTYDPNKRINAEECLHHPWMTKFASHK 64
+ ++++ G + F P+W V+ EAKDL+ KMLT +P+KRI A E L HPW+ SH+
Sbjct 219 HRLYQQIKAGAYDFPSPEWDTVTPEAKDLINKMLTINPSKRITAAEALKHPWI----SHR 274
Query 65 DTSINKHVLTGALGNMKKFQATQKLAQAAILFMGSKLTTVEETKELTTIFRQLDKNGDGQ 124
T + + +KKF A +KL G+ LTT+ T+ + +K DG
Sbjct 275 STVASCMHRQETVDCLKKFNARRKLK-------GAILTTMLATRNFSGGKSGGNKKSDGV 327
Query 125 LDRKELIEGYRKLLDWKGESGEVDEATIEQE 155
RK +L+ ES E TIE E
Sbjct 328 KKRKS--SSSVQLM----ESSESTNTTIEDE 352
> Hs20149547
Length=735
Score = 68.6 bits (166), Expect = 9e-12, Method: Compositional matrix adjust.
Identities = 35/94 (37%), Positives = 52/94 (55%), Gaps = 7/94 (7%)
Query 3 TDQEILKRVERGKFSFEMPDWGHVSEEAKDLVKKMLTYDPNKRINAEECLHHPWMTKF-- 60
T +EIL R+ GKF+ +W VSE AKDLV KML DP++R+ A++ L HPW+T+
Sbjct 621 TPEEILTRIGSGKFTLSGGNWNTVSETAKDLVSKMLHVDPHQRLTAKQVLQHPWVTQKDK 680
Query 61 -----ASHKDTSINKHVLTGALGNMKKFQATQKL 89
SH+D + K + + + T +L
Sbjct 681 LPQSQLSHQDLQLVKGAMAATYSALNSSKPTPQL 714
> Hs14141724
Length=499
Score = 67.4 bits (163), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 40/124 (32%), Positives = 65/124 (52%), Gaps = 22/124 (17%)
Query 5 QEILKRVERGKFSFEMPDWGHVSEEAKDLVKKMLTYDPNKRINAEECLHHPWMTKFAS-- 62
+ ++++ G + F P+W V+ EAKDL+ KMLT +P KRI A E L HPW+ + ++
Sbjct 220 HRLYQQIKAGAYDFPSPEWDTVTPEAKDLINKMLTINPAKRITASEALKHPWICQRSTVA 279
Query 63 ---HKDTSINKHVLTGALGNMKKFQATQKLAQAAILFMGSKLTTVEETKELTTIFRQLDK 119
H+ +++ +KKF A +KL G+ LTT+ T+ + + L K
Sbjct 280 SMMHRQETVD---------CLKKFNARRKLK-------GAILTTMLATRNFSAA-KSLLK 322
Query 120 NGDG 123
DG
Sbjct 323 KPDG 326
> 7304358
Length=493
Score = 67.0 bits (162), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 56/213 (26%), Positives = 95/213 (44%), Gaps = 54/213 (25%)
Query 5 QEILKRVERGKFSFEMPDWGHVSEEAKDLVKKMLTYDPNKRINAEECLHHPWMT---KFA 61
+ +++ G + + P+W V+ EAK+L+ +MLT +PNKRI A E L HPW+ + A
Sbjct 220 HRLYSQIKAGAYDYPSPEWDTVTPEAKNLINQMLTVNPNKRITAAEALKHPWICQRERVA 279
Query 62 S--HKDTSINKHVLTGALGNMKKFQATQKLAQAAILFMGSKLTTVEETKELTTIFRQLDK 119
S H+ +++ +KKF A +KL G+ LTT+ T+ +
Sbjct 280 SVVHRQETVD---------CLKKFNARRKLK-------GAILTTMLATRNFSKF------ 317
Query 120 NGDGQLDRKELIEGYRKLLDWKGESGEVDEATIEQEVDQILATVDFDKNGYIEYSEFVTV 179
G R ++ KGE +V E+T D T++ D +
Sbjct 318 TG-------------RSMITKKGEGSQVKEST-----DSSSTTLEDDD---------IKA 350
Query 180 CMDKQLLLSRERLLQAFQQFDSDGSGKITNDEL 212
++++ E+L++A D DG KI + L
Sbjct 351 ARRQEIIKITEQLIEAINSGDFDGYTKICDPHL 383
> 7300778
Length=450
Score = 65.5 bits (158), Expect = 8e-11, Method: Compositional matrix adjust.
Identities = 54/190 (28%), Positives = 85/190 (44%), Gaps = 28/190 (14%)
Query 11 VERGKFSFEMPDWGHVSEEAKDLVKKMLTYDPNKRINAEECLHHPWMTKFASHKDTSINK 70
V RG+ SFE P+W +S AKDLV KML +P+ R++ E L HPW+ +D
Sbjct 232 VARGRLSFEAPEWKSISANAKDLVMKMLAANPHHRLSITEVLDHPWIRD----RDKLQRT 287
Query 71 HVLTGALGNMKKFQATQKLAQAA-ILFMGSKLTTVEETKELTTIFRQLDKNGDGQLDRKE 129
H L + +K++ A +KL A + G+ + + T I D+ D +
Sbjct 288 H-LADTVEELKRYNARRKLKGAVQAIAGGTNMDPLYATDADMPITGATDEWADEEAG--- 343
Query 130 LIEGYRKLLDWKGESGEVDEATIEQEV-----------------DQILATVDFDKNGYIE 172
IE +++LD + + +A ++ +V D+I ATV NG
Sbjct 344 -IEAVQRILDCLDDIYSLQDAHVDADVLRDMLRDNRLHQFLQLFDRIAATV-VTSNGRAP 401
Query 173 YSEFVTVCMD 182
+E V C D
Sbjct 402 AAEAVGRCRD 411
> CE13014
Length=740
Score = 65.5 bits (158), Expect = 9e-11, Method: Compositional matrix adjust.
Identities = 30/66 (45%), Positives = 42/66 (63%), Gaps = 0/66 (0%)
Query 3 TDQEILKRVERGKFSFEMPDWGHVSEEAKDLVKKMLTYDPNKRINAEECLHHPWMTKFAS 62
T +IL+RV GK S P W +S+EAKDLV+KML DPN+R+ A++ L H W+ + +
Sbjct 601 TPDQILQRVGDGKISMTHPVWDTISDEAKDLVRKMLDVDPNRRVTAKQALQHKWIGQKEA 660
Query 63 HKDTSI 68
D I
Sbjct 661 LPDRPI 666
> CE18169
Length=797
Score = 65.1 bits (157), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 30/66 (45%), Positives = 42/66 (63%), Gaps = 0/66 (0%)
Query 3 TDQEILKRVERGKFSFEMPDWGHVSEEAKDLVKKMLTYDPNKRINAEECLHHPWMTKFAS 62
T +IL+RV GK S P W +S+EAKDLV+KML DPN+R+ A++ L H W+ + +
Sbjct 658 TPDQILQRVGDGKISMTHPVWDTISDEAKDLVRKMLDVDPNRRVTAKQALQHKWIGQKEA 717
Query 63 HKDTSI 68
D I
Sbjct 718 LPDRPI 723
> Hs4502567_1
Length=360
Score = 65.1 bits (157), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 33/97 (34%), Positives = 53/97 (54%), Gaps = 4/97 (4%)
Query 3 TDQEILKRVERGKFSFEMPDWGHVSEEAKDLVKKMLTYDPNKRINAEECLHHPWMTKFAS 62
T + + + + +GK+ W H+SE AKDLV++ML DP +RI E L+HPW+ +
Sbjct 222 TKERLFEGIIKGKYKMNPRQWSHISESAKDLVRRMLMLDPAERITVYEALNHPWLKE--- 278
Query 63 HKDTSINKHVLTGALGNMKKFQATQKLAQAAILFMGS 99
+D K L + ++KF A +KL A + + S
Sbjct 279 -RDRYAYKIHLPETVEQLRKFNARRKLKGAVLAAVSS 314
> Hs4759050
Length=740
Score = 64.7 bits (156), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 32/80 (40%), Positives = 45/80 (56%), Gaps = 4/80 (5%)
Query 3 TDQEILKRVERGKFSFEMPDWGHVSEEAKDLVKKMLTYDPNKRINAEECLHHPWMTKFAS 62
T +EIL R+ GKFS W VS+ AKDLV KML DP++R+ A L HPW+ +
Sbjct 625 TPEEILARIGSGKFSLSGGYWNSVSDTAKDLVSKMLHVDPHQRLTAALVLRHPWIVHWDQ 684
Query 63 HKDTSINK----HVLTGALG 78
+N+ H++ GA+
Sbjct 685 LPQYQLNRQDAPHLVKGAMA 704
> Hs20127418
Length=664
Score = 63.9 bits (154), Expect = 2e-10, Method: Composition-based stats.
Identities = 34/115 (29%), Positives = 61/115 (53%), Gaps = 21/115 (18%)
Query 2 QTDQEILKRVERGKFSFEMPDWGHVSEEAKDLVKKMLTYDPNKRINAEECLHHPWMTKFA 61
+ ++ ++++ G + F P+W V+ EAK+L+ +MLT +P KRI A E L HPW+ + +
Sbjct 217 EDQHKLYQQIKAGAYDFPSPEWDTVTPEAKNLINQMLTINPAKRITAHEALKHPWVCQRS 276
Query 62 S-----HKDTSINKHVLTGALGNMKKFQATQKLAQAAILFMGSKLTTVEETKELT 111
+ H+ ++ +KKF A +KL G+ LTT+ T+ +
Sbjct 277 TVASMMHRQETVE---------CLKKFNARRKLK-------GAILTTMLATRNFS 315
> Hs19923570
Length=733
Score = 62.8 bits (151), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 36/115 (31%), Positives = 61/115 (53%), Gaps = 7/115 (6%)
Query 3 TDQEILKRVERGKFSFEMPDWGHVSEEAKDLVKKMLTYDPNKRINAEECLHHPWMTK--- 59
T +EIL R+ GK++ +W +S+ AKD+V KML DP++R+ A + L HPW+
Sbjct 618 TPEEILARIGSGKYALSGGNWDSISDAAKDVVSKMLHVDPHQRLTAMQVLKHPWVVNREY 677
Query 60 FASHKDTSINKHVLTGALGNMKKFQATQKLAQAAIL--FMGSKLTTVEETKELTT 112
+ ++ + + H++ GA+ + A + QA L + S L K LT+
Sbjct 678 LSPNQLSRQDVHLVKGAMA--ATYFALNRTPQAPRLEPVLSSNLAQRRGMKRLTS 730
> Hs7657526
Length=745
Score = 61.2 bits (147), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 33/83 (39%), Positives = 47/83 (56%), Gaps = 12/83 (14%)
Query 3 TDQEILKRVERGKFSFEMPDWGHVSEEAKDLVKKMLTYDPNKRINAEECLHHPWMTKFAS 62
T +EIL R+ GKFS +W ++S+ AKDL+ ML DP++R AE+ L H W+T
Sbjct 629 TPEEILLRIGNGKFSLSGGNWDNISDGAKDLLSHMLHMDPHQRYTAEQILKHSWIT---- 684
Query 63 HKDTSIN--------KHVLTGAL 77
H+D N HV+ GA+
Sbjct 685 HRDQLPNDQPKRNDVSHVVKGAM 707
> Hs9966875
Length=357
Score = 61.2 bits (147), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 26/91 (28%), Positives = 50/91 (54%), Gaps = 5/91 (5%)
Query 2 QTDQEILKRVERGKFSFEMPDWGHVSEEAKDLVKKMLTYDPNKRINAEECLHHPWMTKFA 61
+ D ++ +++ + ++ F+ P W +S+ AKD ++ ++ DPNKR E+ HPW+
Sbjct 224 ENDSKLFEQILKAEYEFDSPYWDDISDSAKDFIRNLMEKDPNKRYTCEQAARHPWIA--- 280
Query 62 SHKDTSINKHVLTGALGNMKKFQATQKLAQA 92
DT++NK++ ++K A K QA
Sbjct 281 --GDTALNKNIHESVSAQIRKNFAKSKWRQA 309
> 7304283
Length=7175
Score = 60.8 bits (146), Expect = 2e-09, Method: Composition-based stats.
Identities = 27/70 (38%), Positives = 42/70 (60%), Gaps = 0/70 (0%)
Query 1 GQTDQEILKRVERGKFSFEMPDWGHVSEEAKDLVKKMLTYDPNKRINAEECLHHPWMTKF 60
G D + LK V+ + F++ + ++SEEAKD ++K+L + KR+ A ECL HPW+T
Sbjct 6337 GDNDVQTLKNVKACDWDFDVESFKYISEEAKDFIRKLLVRNKEKRMTAHECLLHPWLTGD 6396
Query 61 ASHKDTSINK 70
S IN+
Sbjct 6397 HSAMKQEINR 6406
> CE28057
Length=520
Score = 59.7 bits (143), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 35/118 (29%), Positives = 59/118 (50%), Gaps = 21/118 (17%)
Query 5 QEILKRVERGKFSFEMPDWGHVSEEAKDLVKKMLTYDPNKRINAEECLHHPWMTK----- 59
+ +++ G + + P+W V+ EAK L+ MLT +P KRI A++ L PW+
Sbjct 217 HRLYAQIKAGAYDYPSPEWDTVTPEAKSLIDSMLTVNPKKRITADQALKVPWICNRERVA 276
Query 60 FASHKDTSINKHVLTGALGNMKKFQATQKLAQAAILFMGSKLTTVEETKELTTIFRQL 117
A H+ +++ +KKF A +KL G+ LTT+ T+ L+ + R L
Sbjct 277 SAIHRQDTVD---------CLKKFNARRKLK-------GAILTTMIATRNLSNLGRNL 318
> CE28059
Length=350
Score = 59.7 bits (143), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 35/118 (29%), Positives = 59/118 (50%), Gaps = 21/118 (17%)
Query 5 QEILKRVERGKFSFEMPDWGHVSEEAKDLVKKMLTYDPNKRINAEECLHHPWMTK----- 59
+ +++ G + + P+W V+ EAK L+ MLT +P KRI A++ L PW+
Sbjct 217 HRLYAQIKAGAYDYPSPEWDTVTPEAKSLIDSMLTVNPKKRITADQALKVPWICNRERVA 276
Query 60 FASHKDTSINKHVLTGALGNMKKFQATQKLAQAAILFMGSKLTTVEETKELTTIFRQL 117
A H+ +++ +KKF A +KL G+ LTT+ T+ L+ + R L
Sbjct 277 SAIHRQDTVD---------CLKKFNARRKLK-------GAILTTMIATRNLSNLGRNL 318
Lambda K H
0.316 0.133 0.379
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4147789496
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40