bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0630_orf2
Length=155
Score E
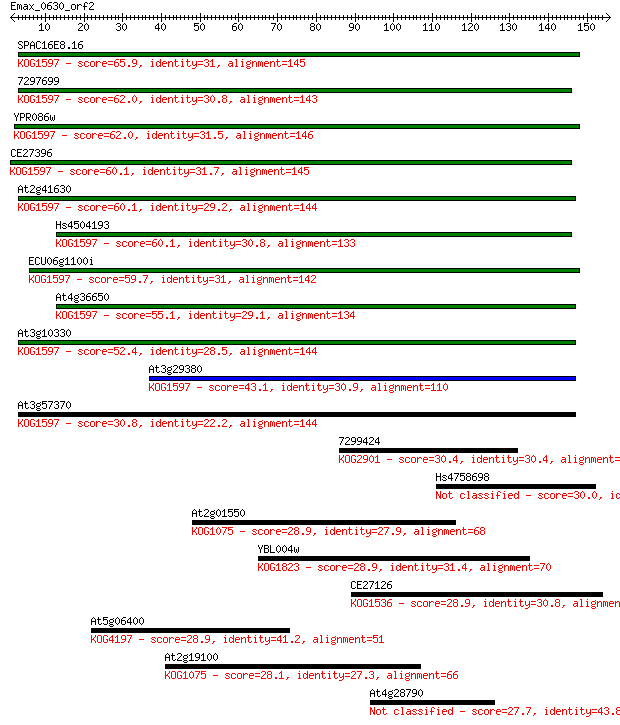
Sequences producing significant alignments: (Bits) Value
SPAC16E8.16 65.9 3e-11
7297699 62.0 4e-10
YPR086w 62.0 4e-10
CE27396 60.1 2e-09
At2g41630 60.1 2e-09
Hs4504193 60.1 2e-09
ECU06g1100i 59.7 2e-09
At4g36650 55.1 5e-08
At3g10330 52.4 3e-07
At3g29380 43.1 2e-04
At3g57370 30.8 1.2
7299424 30.4 1.4
Hs4758698 30.0 1.7
At2g01550 28.9 3.8
YBL004w 28.9 3.8
CE27126 28.9 4.4
At5g06400 28.9 4.5
At2g19100 28.1 7.2
At4g28790 27.7 9.5
> SPAC16E8.16
Length=340
Score = 65.9 bits (159), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 45/160 (28%), Positives = 82/160 (51%), Gaps = 16/160 (10%)
Query 3 IAEAFCLRDYILERAKEIAKELLDAGQLRSRSTCVCMLAITYLACREGGATRTVKELIVY 62
+ +A L I + AK++ K + D L+ +S+ + A Y+ACR+G RT E+
Sbjct 128 MCDAISLPKVIADTAKQLYKRVDDHKALKGKSSQSIIAACIYIACRQGKVPRTFMEICTL 187
Query 63 DRSIKEKELGKQINKIKRLLPNRGGGNN-------QENVQ-------QLLPRYCSKLQLS 108
++ +KE+G+ ++R+L G +N E +Q L+ R+C++L L
Sbjct 188 -TNVPKKEIGRVYKTLQRMLTEGGALHNSVDALKGHEYIQSSSTSAEDLMVRFCNRLMLP 246
Query 109 MHVSDVAEYVAKRAA-QVFVSSHRPNSVAAAAIWLVVQLL 147
M V A +A+RA Q ++ P S+AA+ I+++ L+
Sbjct 247 MSVQSAAAELARRAGLQGTLAGRSPISIAASGIYMISALM 286
> 7297699
Length=315
Score = 62.0 bits (149), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 44/144 (30%), Positives = 71/144 (49%), Gaps = 5/144 (3%)
Query 3 IAEAFCLRDYILERAKEIAKELLDAGQLRSRSTCVCMLAITYLACREGGATRTVKELIVY 62
+A+ L I++RA + K++ D L+ RS A Y+ACR+ G RT KE+
Sbjct 123 MADRINLPKTIVDRANNLFKQVHDGKNLKGRSNDAKASACLYIACRQEGVPRTFKEICAV 182
Query 63 DRSIKEKELGKQINKIKRLLPNRGGGNNQENVQQLLPRYCSKLQLSMHVSDVAEYVAKRA 122
+ I +KE+G+ + L + + R+C+ L L V A ++AK+A
Sbjct 183 SK-ISKKEIGRCFKLTLKALET---SVDLITTADFMCRFCANLDLPNMVQRAATHIAKKA 238
Query 123 AQV-FVSSHRPNSVAAAAIWLVVQ 145
++ V P SVAAAAI++ Q
Sbjct 239 VEMDIVPGRSPISVAAAAIYMASQ 262
> YPR086w
Length=345
Score = 62.0 bits (149), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 46/158 (29%), Positives = 77/158 (48%), Gaps = 15/158 (9%)
Query 2 LIAEAFCLRDYILERAKEIAKELLDAGQLRSRSTCVCMLAITYLACREGGATRTVKEL-- 59
++ +A L + + AKE K D L+ +S M A + CR RT KE+
Sbjct 135 MLCDAAELPKIVKDCAKEAYKLCHDEKTLKGKSMESIMAASILIGCRRAEVARTFKEIQS 194
Query 60 IVYDRSIKEKELGKQINKIKRLLPNRG-------GGNNQENVQQL--LPRYCSKLQLSMH 110
+++ +K KE GK +N +K +L + +N Q L +PR+CS L L M
Sbjct 195 LIH---VKTKEFGKTLNIMKNILRGKSEDGFLKIDTDNMSGAQNLTYIPRFCSHLGLPMQ 251
Query 111 VSDVAEYVAKRAAQVF-VSSHRPNSVAAAAIWLVVQLL 147
V+ AEY AK+ ++ ++ P ++A +I+L + L
Sbjct 252 VTTSAEYTAKKCKEIKEIAGKSPITIAVVSIYLNILLF 289
> CE27396
Length=306
Score = 60.1 bits (144), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 46/151 (30%), Positives = 75/151 (49%), Gaps = 15/151 (9%)
Query 1 RLIAEAFCLRDYILERAKEIAKELLDAGQLRSRSTCVCMLAITYLACREGGATRTVKELI 60
R ++E L I + A I K++L++ LR ++ A Y+ACR+ G RT KE+
Sbjct 112 REMSERIHLPRNIQDSASRIFKDVLESKALRGKNNEAQAAACLYIACRKDGVPRTFKEIC 171
Query 61 VYDRSIKEKELGKQINKIKRLLPNRGGGNNQENVQQL-----LPRYCSKLQLSMHVSDVA 115
R + +KE+G+ I R L + N++Q+ + R+C L L + A
Sbjct 172 AVSR-VSKKEIGRCFKIIVRSL--------ETNLEQITSADFMSRFCGNLSLPNSIQAAA 222
Query 116 EYVAKRAAQV-FVSSHRPNSVAAAAIWLVVQ 145
+AK A + V+ P S+AAAAI++ Q
Sbjct 223 TRIAKCAVDMDLVAGRTPISIAAAAIYMASQ 253
> At2g41630
Length=312
Score = 60.1 bits (144), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 42/147 (28%), Positives = 70/147 (47%), Gaps = 4/147 (2%)
Query 3 IAEAFCLRDYILERAKEIAKELLDAGQLRSRSTCVCMLAITYLACREGGATRTVKELIVY 62
++E L I +RA E+ K L D R R+ A Y+ACR+ RT+KE+ V
Sbjct 115 MSERLGLVATIKDRANELYKRLEDQKSSRGRNQDALYAACLYIACRQEDKPRTIKEICVI 174
Query 63 DRSIKEKELGKQINKIKRLL---PNRGGGNNQENVQQLLPRYCSKLQLSMHVSDVAEYVA 119
+KE+G+ + I + L P + + + R+CS L +S H A+
Sbjct 175 ANGATKKEIGRAKDYIVKTLGLEPGQSVDLGTIHAGDFMRRFCSNLAMSNHAVKAAQEAV 234
Query 120 KRAAQVFVSSHRPNSVAAAAIWLVVQL 146
+++ + F P S+AA I+++ QL
Sbjct 235 QKSEE-FDIRRSPISIAAVVIYIITQL 260
> Hs4504193
Length=316
Score = 60.1 bits (144), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 41/134 (30%), Positives = 67/134 (50%), Gaps = 5/134 (3%)
Query 13 ILERAKEIAKELLDAGQLRSRSTCVCMLAITYLACREGGATRTVKELIVYDRSIKEKELG 72
I++R + K++ + L+ R+ A Y+ACR+ G RT KE+ R I +KE+G
Sbjct 134 IVDRTNNLFKQVYEQKSLKGRANDAIASACLYIACRQEGVPRTFKEICAVSR-ISKKEIG 192
Query 73 KQINKIKRLLPNRGGGNNQENVQQLLPRYCSKLQLSMHVSDVAEYVAKRAAQV-FVSSHR 131
+ I + L + + R+CS L L V A ++A++A ++ V
Sbjct 193 RCFKLILKALET---SVDLITTGDFMSRFCSNLCLPKQVQMAATHIARKAVELDLVPGRS 249
Query 132 PNSVAAAAIWLVVQ 145
P SVAAAAI++ Q
Sbjct 250 PISVAAAAIYMASQ 263
> ECU06g1100i
Length=312
Score = 59.7 bits (143), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 44/147 (29%), Positives = 79/147 (53%), Gaps = 10/147 (6%)
Query 6 AFCLRD----YILERAKEIAKELLDAGQLRSRSTCVCMLAITYLACREGGATRTVKELIV 61
AFC R I++RA I K + + L+ ++ + A Y+ACR+ RT KE+ V
Sbjct 119 AFCERSNLSRTIIDRAHYIFKNIEERKLLKGKNVEGIVAACIYIACRQEECPRTFKEISV 178
Query 62 YDRSIKEKELGKQINKIKRLLPNRGGGNNQENVQQLLPRYCSKLQLSMHVSDVAEYVAKR 121
+++++E+G+ I L R + EN+ + R+CS L L++ + +A +AK
Sbjct 179 M-TAVQKREIGRCFKLISPHL-ERMATMSTENI---IARFCSDLNLNIKIQKIATEIAKA 233
Query 122 AAQV-FVSSHRPNSVAAAAIWLVVQLL 147
A ++ ++ P+S+AAA I++V L
Sbjct 234 AHELGCLAGKSPDSIAAAVIYMVTNLF 260
> At4g36650
Length=503
Score = 55.1 bits (131), Expect = 5e-08, Method: Composition-based stats.
Identities = 39/135 (28%), Positives = 70/135 (51%), Gaps = 3/135 (2%)
Query 13 ILERAKEIAKELLDAGQLRSRSTCVCMLAITYLACREGGATRTVKELIVYDRSIKEKELG 72
I E A ++ ++ A LR+RS A A RE RT++E+ + ++++KE+G
Sbjct 150 ISEHAFQLFRDCCSATCLRNRSVEALATACLVQAIREAQEPRTLQEISIA-ANVQQKEIG 208
Query 73 KQINKIKRLLPNRGGGNNQENVQQLLPRYCSKLQLSMHVSDVAEYVAKRAA-QVFVSSHR 131
K I + L N ++ +PR+C+ LQL+ ++A ++ + + F +
Sbjct 209 KYIKILGEAL-QLSQPINSNSISVHMPRFCTLLQLNKSAQELATHIGEVVINKCFCTRRN 267
Query 132 PNSVAAAAIWLVVQL 146
P S++AAAI+L QL
Sbjct 268 PISISAAAIYLACQL 282
> At3g10330
Length=312
Score = 52.4 bits (124), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 41/147 (27%), Positives = 68/147 (46%), Gaps = 4/147 (2%)
Query 3 IAEAFCLRDYILERAKEIAKELLDAGQLRSRSTCVCMLAITYLACREGGATRTVKELIVY 62
+A+ L I +RA EI K + D R R+ + A Y+ACR+ RTVKE+
Sbjct 115 MADRLGLVATIKDRANEIYKRVEDQKSSRGRNQDALLAACLYIACRQEDKPRTVKEICSV 174
Query 63 DRSIKEKELGKQINKIKRLLPNRGGG---NNQENVQQLLPRYCSKLQLSMHVSDVAEYVA 119
+KE+G+ I + L G + + R+CS L ++ A+
Sbjct 175 ANGATKKEIGRAKEYIVKQLGLETGQLVEMGTIHAGDFMRRFCSNLGMTNQTVKAAQESV 234
Query 120 KRAAQVFVSSHRPNSVAAAAIWLVVQL 146
+++ + F P S+AAA I+++ QL
Sbjct 235 QKSEE-FDIRRSPISIAAAVIYIITQL 260
> At3g29380
Length=336
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 34/115 (29%), Positives = 60/115 (52%), Gaps = 8/115 (6%)
Query 37 VCMLAITYLACREGGATRTVKELIVYDRSIKEKELGKQINKIKRLLPNRGGGNNQE---- 92
+C +++ ACRE +RT+KE+ + +K++ K+ IKR+L + +
Sbjct 160 ICAASVS-TACRELQLSRTLKEIAEVANGVDKKDIRKESLVIKRVLESHQTSVSASQAII 218
Query 93 NVQQLLPRYCSKLQLSM-HVSDVAEYVAKRAAQVFVSSHRPNSVAAAAIWLVVQL 146
N +L+ R+CSKL +S + + E V K A+ F P SV AA I+++ +
Sbjct 219 NTGELVRRFCSKLDISQREIMAIPEAVEK--AENFDIRRNPKSVLAAIIFMISHI 271
> At3g57370
Length=360
Score = 30.8 bits (68), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 32/144 (22%), Positives = 59/144 (40%), Gaps = 2/144 (1%)
Query 3 IAEAFCLRDYILERAKEIAKELLDAGQLRSRSTCVCMLAITYLACREGGATRTVKELIVY 62
I+ L I +A EI K + + + R+ V A Y+ACR+ TRT++E+ +
Sbjct 167 ISNGLKLPATIKGQANEIFKVVESYARGKERN--VLFAACIYIACRDNDMTRTMREISRF 224
Query 63 DRSIKEKELGKQINKIKRLLPNRGGGNNQENVQQLLPRYCSKLQLSMHVSDVAEYVAKRA 122
++ + + I L + R+CS +L + A A+
Sbjct 225 ANKASISDISETVGFIAEKLEINKNWYMSIETANFIKRFCSIFRLDKEAVEAALEAAESY 284
Query 123 AQVFVSSHRPNSVAAAAIWLVVQL 146
+ P SVAA ++++ +L
Sbjct 285 DYMTNGRRAPVSVAAGIVYVIARL 308
> 7299424
Length=437
Score = 30.4 bits (67), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 14/46 (30%), Positives = 27/46 (58%), Gaps = 0/46 (0%)
Query 86 GGGNNQENVQQLLPRYCSKLQLSMHVSDVAEYVAKRAAQVFVSSHR 131
G G +V ++L ++ + SMH+ +V+ +++K AQ F SH+
Sbjct 135 GRGTLARDVLKVLTKFKQDAEFSMHMVEVSPFLSKAQAQRFCYSHQ 180
> Hs4758698
Length=1143
Score = 30.0 bits (66), Expect = 1.7, Method: Composition-based stats.
Identities = 13/41 (31%), Positives = 23/41 (56%), Gaps = 0/41 (0%)
Query 111 VSDVAEYVAKRAAQVFVSSHRPNSVAAAAIWLVVQLLSSSS 151
+S V+ YV+ QVF +S +P V +A+W +S ++
Sbjct 661 ISLVSVYVSSPTVQVFSASGKPVEVQVSAVWDTANTISETA 701
> At2g01550
Length=1449
Score = 28.9 bits (63), Expect = 3.8, Method: Composition-based stats.
Identities = 19/68 (27%), Positives = 32/68 (47%), Gaps = 7/68 (10%)
Query 48 REGGATRTVKELIVYDRSIKEKELGKQINKIKRLLPNRGGGNNQENVQQLLPRYCSKLQL 107
+E G+T T K+ I + E + + +L+PN G E + LLP +CS +
Sbjct 809 KEDGSTATTKDDI-------KNETERFFQEFLQLIPNDYEGITVEKLTSLLPYHCSPAEK 861
Query 108 SMHVSDVA 115
M + V+
Sbjct 862 DMLTASVS 869
> YBL004w
Length=2493
Score = 28.9 bits (63), Expect = 3.8, Method: Composition-based stats.
Identities = 22/70 (31%), Positives = 33/70 (47%), Gaps = 2/70 (2%)
Query 65 SIKEKELGKQINKIKRLLPNRGGGNNQENVQQLLPRYCSKLQLSMHVSDVAEYVAKRAAQ 124
+I E G ++ +K LL R NQ + +LL RY L L+ + +E + K Q
Sbjct 1760 NISLTEFGTLLSPVKALLMVRINLRNQNKLSELLRRYL--LGLNHNSDSESESILKFCHQ 1817
Query 125 VFVSSHRPNS 134
+F S NS
Sbjct 1818 LFQESEMSNS 1827
> CE27126
Length=1025
Score = 28.9 bits (63), Expect = 4.4, Method: Composition-based stats.
Identities = 20/69 (28%), Positives = 33/69 (47%), Gaps = 4/69 (5%)
Query 89 NNQENVQQLLPRYCSKLQLSMHVSDVAEYVAKRAAQVFVSSHRPNSVAAAAIWL----VV 144
NN++N Q+ + ++ VS EY R+ + F S+ P+ VA + +
Sbjct 396 NNRQNSQRRFSAFSTESDRGRSVSPSFEYYQNRSLRGFTSAANPHRVATRNSTVRQVPLY 455
Query 145 QLLSSSSSS 153
Q L+S SSS
Sbjct 456 QFLTSRSSS 464
> At5g06400
Length=1030
Score = 28.9 bits (63), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 21/60 (35%), Positives = 31/60 (51%), Gaps = 12/60 (20%)
Query 22 KELLDAGQLRSRSTCVCMLAITYLACREGG-----ATRTVKELI----VYDRSIKEKELG 72
KE+ D G + S ST C++ + C + G ATRT +E+I V DR + + LG
Sbjct 737 KEMKDMGLIPSSSTFKCLITVL---CEKKGRNVEEATRTFREMIRSGFVPDRELVQDYLG 793
> At2g19100
Length=1447
Score = 28.1 bits (61), Expect = 7.2, Method: Composition-based stats.
Identities = 18/67 (26%), Positives = 32/67 (47%), Gaps = 10/67 (14%)
Query 41 AITYLACREGGATRTVKELIVY-DRSIKEKELGKQINKIKRLLPNRGGGNNQENVQQLLP 99
+I+ + C++G T E+ Y +R +E +L+PN G ++Q LLP
Sbjct 796 SISEIQCQDGSVTAKGDEIKAYAERFFRE---------FLQLIPNEYEGVTMADLQDLLP 846
Query 100 RYCSKLQ 106
CS+ +
Sbjct 847 FRCSETE 853
> At4g28790
Length=413
Score = 27.7 bits (60), Expect = 9.5, Method: Compositional matrix adjust.
Identities = 14/32 (43%), Positives = 20/32 (62%), Gaps = 1/32 (3%)
Query 94 VQQLLPRYCSKLQLSMHVSDVAEYVAKRAAQV 125
+Q+LLPR C+K S + DV EYV +Q+
Sbjct 300 LQELLPR-CTKTDRSSMLDDVIEYVKSLQSQI 330
Lambda K H
0.319 0.131 0.364
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2033998736
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40