bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0653_orf2
Length=173
Score E
Sequences producing significant alignments: (Bits) Value
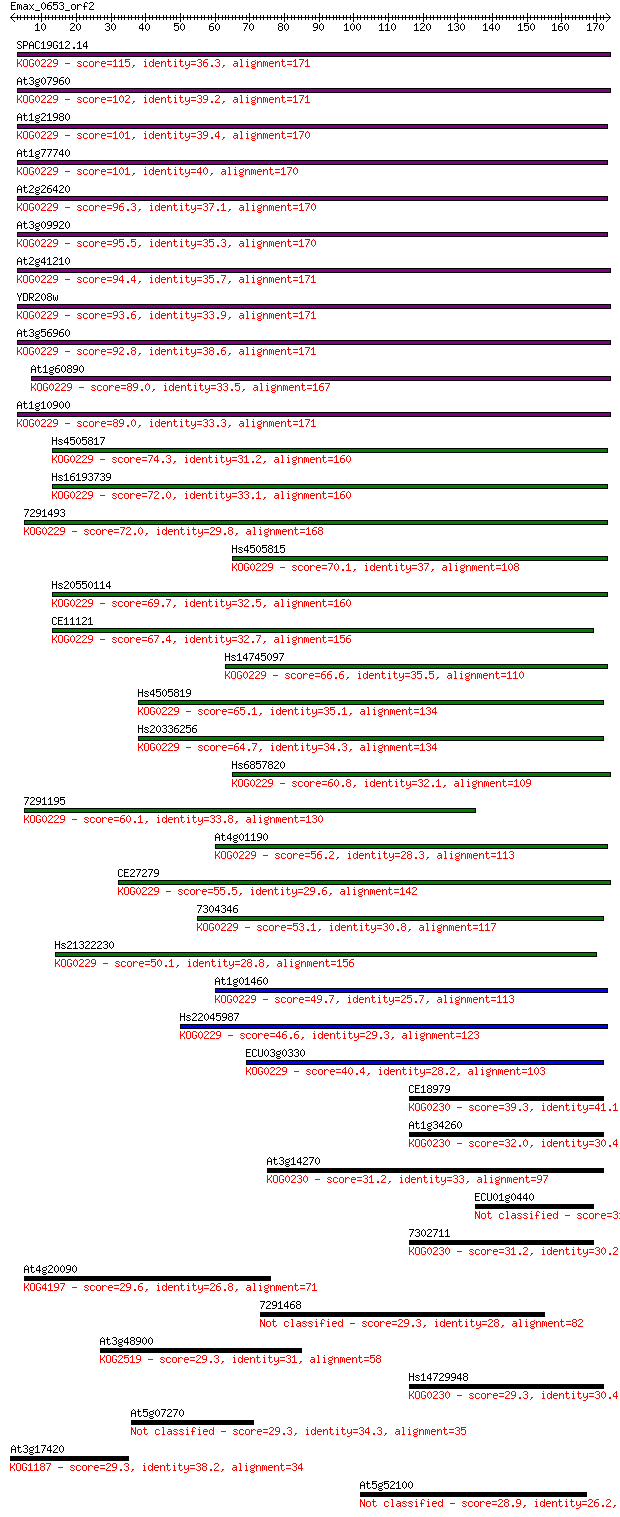
SPAC19G12.14 115 4e-26
At3g07960 102 3e-22
At1g21980 101 6e-22
At1g77740 101 9e-22
At2g26420 96.3 2e-20
At3g09920 95.5 5e-20
At2g41210 94.4 9e-20
YDR208w 93.6 2e-19
At3g56960 92.8 3e-19
At1g60890 89.0 4e-18
At1g10900 89.0 4e-18
Hs4505817 74.3 1e-13
Hs16193739 72.0 6e-13
7291493 72.0 6e-13
Hs4505815 70.1 2e-12
Hs20550114 69.7 3e-12
CE11121 67.4 1e-11
Hs14745097 66.6 2e-11
Hs4505819 65.1 8e-11
Hs20336256 64.7 8e-11
Hs6857820 60.8 1e-09
7291195 60.1 2e-09
At4g01190 56.2 3e-08
CE27279 55.5 6e-08
7304346 53.1 3e-07
Hs21322230 50.1 2e-06
At1g01460 49.7 3e-06
Hs22045987 46.6 3e-05
ECU03g0330 40.4 0.002
CE18979 39.3 0.004
At1g34260 32.0 0.60
At3g14270 31.2 0.95
ECU01g0440 31.2 1.2
7302711 31.2 1.2
At4g20090 29.6 3.4
7291468 29.3 3.7
At3g48900 29.3 3.7
Hs14729948 29.3 4.0
At5g07270 29.3 4.2
At3g17420 29.3 4.2
At5g52100 28.9 5.5
> SPAC19G12.14
Length=742
Score = 115 bits (288), Expect = 4e-26, Method: Composition-based stats.
Identities = 62/171 (36%), Positives = 95/171 (55%), Gaps = 15/171 (8%)
Query 3 GHESWNMVINIMVGIRLAGARAMSEPHRAVEPYDFLMKEKFSVLPKTGMVDKKRRKPSLC 62
GHE++ N++ GIR+ +R ++ R + P DF + KF+ + + +
Sbjct 264 GHENYVTAYNMLTGIRVGVSRCQAKMDRELTPADFTARHKFTF----DITGNELTPSAKY 319
Query 63 AVRFIDYAPMVFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSLSELVSEGKSGSLF 122
+F DYAP VFR LR LF +D+ Y+ S+ +L SEL S GKSGS F
Sbjct 320 DFKFKDYAPWVFRHLRQLFHLDAADYLVSLTSKYIL-----------SELDSPGKSGSFF 368
Query 123 YYTTDGRFMIKTVSKETAFFMRSILFDYYKHVSTCTNTMLTRLCGLHALRL 173
Y++ D RF+IKT+ F+R IL+DYY+HV NT++++ GLH ++L
Sbjct 369 YFSRDYRFIIKTIHHSEHKFLREILYDYYEHVKNNPNTLISQFYGLHRVKL 419
> At3g07960
Length=715
Score = 102 bits (255), Expect = 3e-22, Method: Compositional matrix adjust.
Identities = 67/174 (38%), Positives = 96/174 (55%), Gaps = 18/174 (10%)
Query 3 GHESWNMVINIMVGIRLAGARAMSEPHRAVEPYDFLMKEK-FSVLPKTGMVDKKRRKP-S 60
GH+++ +++N+ +GIR + R ++ F KEK ++ P G K P
Sbjct 325 GHKNYELMLNLQLGIRHSVGRPAPATSLDLKASAFDPKEKLWTKFPSEG---SKYTPPHQ 381
Query 61 LCAVRFIDYAPMVFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSLSELVSEGKSGS 120
C ++ DY P+VFR LR LF +D+ Y+ S I GN +L EL S GKSGS
Sbjct 382 SCEFKWKDYCPVVFRTLRKLFSVDAADYMLS----------ICGN-DALRELSSPGKSGS 430
Query 121 LFYYTTDGRFMIKTVSK-ETAFFMRSILFDYYKHVSTCTNTMLTRLCGLHALRL 173
FY T D R+MIKT+ K ET +R +L YY HV C NT++T+ GLH ++L
Sbjct 431 FFYLTNDDRYMIKTMKKAETKVLIR-MLPAYYNHVRACENTLVTKFFGLHCVKL 483
> At1g21980
Length=752
Score = 101 bits (252), Expect = 6e-22, Method: Compositional matrix adjust.
Identities = 67/173 (38%), Positives = 96/173 (55%), Gaps = 19/173 (10%)
Query 3 GHESWNMVINIMVGIRLAGARAMSEPHRAVEPYDFLMKEKF-SVLPKTGMVDKKRRKPSL 61
GH+ +++++N+ +GIR + + S R ++ DF KEKF + P G + P
Sbjct 351 GHKKYDLMLNLQLGIRYSVGKHAS-IVRDLKQTDFDPKEKFWTRFPPEGT----KTTPPH 405
Query 62 CAV--RFIDYAPMVFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSLSELVSEGKSG 119
+V R+ DY P+VFRRLR LF +D Y+ + I GN +L EL S GKSG
Sbjct 406 QSVDFRWKDYCPLVFRRLRELFQVDPAKYMLA----------ICGN-DALRELSSPGKSG 454
Query 120 SLFYYTTDGRFMIKTVSKETAFFMRSILFDYYKHVSTCTNTMLTRLCGLHALR 172
S FY T D RFMIKTV K + +L YYKHV N+++TR G+H ++
Sbjct 455 SFFYLTQDDRFMIKTVKKSEVKVLLRMLPSYYKHVCQYENSLVTRFYGVHCVK 507
> At1g77740
Length=754
Score = 101 bits (251), Expect = 9e-22, Method: Compositional matrix adjust.
Identities = 68/172 (39%), Positives = 93/172 (54%), Gaps = 17/172 (9%)
Query 3 GHESWNMVINIMVGIRLAGARAMSEPHRAVEPYDFLMKEKF-SVLPKTGMVDKKRRKPSL 61
GH+ +++++N+ GIR + + S R ++ DF EKF + P G K P L
Sbjct 353 GHKKYDLMLNLQHGIRYSVGKHAS-VVRDLKQSDFDPSEKFWTRFPPEG---SKTTPPHL 408
Query 62 CA-VRFIDYAPMVFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSLSELVSEGKSGS 120
R+ DY P+VFRRLR LF +D Y+ + I GN +L EL S GKSGS
Sbjct 409 SVDFRWKDYCPLVFRRLRELFTVDPADYMLA----------ICGN-DALRELSSPGKSGS 457
Query 121 LFYYTTDGRFMIKTVSKETAFFMRSILFDYYKHVSTCTNTMLTRLCGLHALR 172
FY T D RFMIKTV K + +L YYKHV NT++TR G+H ++
Sbjct 458 FFYLTQDDRFMIKTVKKSEVKVLLRMLPSYYKHVCQYENTLVTRFYGVHCIK 509
> At2g26420
Length=705
Score = 96.3 bits (238), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 63/172 (36%), Positives = 96/172 (55%), Gaps = 17/172 (9%)
Query 3 GHESWNMVINIMVGIRLAGARAMSEPHRAVEPYDFLMKEK-FSVLPKTGMVDKKRRKPSL 61
GH+++++++N+ +GIR + + S R + DF K+K ++ P G K P L
Sbjct 325 GHKNYDLMLNLQLGIRYSVGKHAS-LLRELRHSDFDPKDKQWTRFPPEG---SKSTPPHL 380
Query 62 CA-VRFIDYAPMVFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSLSELVSEGKSGS 120
A ++ DY P+VFR LR LF ID Y+ + I GN SL E S GKSGS
Sbjct 381 SAEFKWKDYCPIVFRHLRDLFAIDQADYMLA----------ICGN-ESLREFASPGKSGS 429
Query 121 LFYYTTDGRFMIKTVSKETAFFMRSILFDYYKHVSTCTNTMLTRLCGLHALR 172
FY T D R+MIKT+ K + +L +YY+HVS N+++T+ G+H ++
Sbjct 430 AFYLTQDERYMIKTMKKSEIKVLLKMLPNYYEHVSKYKNSLVTKFFGVHCVK 481
> At3g09920
Length=815
Score = 95.5 bits (236), Expect = 5e-20, Method: Compositional matrix adjust.
Identities = 60/171 (35%), Positives = 90/171 (52%), Gaps = 14/171 (8%)
Query 3 GHESWNMVINIMVGIRLAGARAMSEPHRAVEPYDFLMKEKFSV-LPKTGMVDKKRRKPSL 61
GH S+++++++ +GIR + R V DF + F + P+ G
Sbjct 391 GHRSYDLMLSLQLGIRYTVGKITPIQRRQVRTADFGPRASFWMTFPRAGSTMTPPHHSE- 449
Query 62 CAVRFIDYAPMVFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSLSELVSEGKSGSL 121
++ DY PMVFR LR +F ID+ Y+ S I GN +L EL S GKSGS+
Sbjct 450 -DFKWKDYCPMVFRNLREMFKIDAADYMMS----------ICGN-DTLRELSSPGKSGSV 497
Query 122 FYYTTDGRFMIKTVSKETAFFMRSILFDYYKHVSTCTNTMLTRLCGLHALR 172
F+ + D RFMIKT+ K + +L DY+ HV T NT++T+ GLH ++
Sbjct 498 FFLSQDDRFMIKTLRKSEVKVLLRMLPDYHHHVKTYENTLITKFFGLHRIK 548
> At2g41210
Length=772
Score = 94.4 bits (233), Expect = 9e-20, Method: Compositional matrix adjust.
Identities = 61/173 (35%), Positives = 89/173 (51%), Gaps = 16/173 (9%)
Query 3 GHESWNMVINIMVGIRLAGARAMSEPHRAVEPYDFLMKEK-FSVLPKTGMVDKKRRKP-S 60
GH ++ +++N+ +GIR + R ++P F K+K + P+ G K P
Sbjct 379 GHRNYELMLNLQLGIRHSVGRQAPAASLDLKPSAFDPKDKIWRRFPREGT---KYTPPHQ 435
Query 61 LCAVRFIDYAPMVFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSLSELVSEGKSGS 120
++ DY P+VFR LR LF +D Y+ S I GN +L EL S GKSGS
Sbjct 436 STEFKWKDYCPLVFRSLRKLFKVDPADYMLS----------ICGN-DALRELSSPGKSGS 484
Query 121 LFYYTTDGRFMIKTVSKETAFFMRSILFDYYKHVSTCTNTMLTRLCGLHALRL 173
FY T D R+MIKT+ K + +L YY HV N+++ R GLH ++L
Sbjct 485 FFYLTNDDRYMIKTMKKSETKVLLGMLAAYYNHVRAFENSLVIRFFGLHCVKL 537
> YDR208w
Length=779
Score = 93.6 bits (231), Expect = 2e-19, Method: Composition-based stats.
Identities = 58/171 (33%), Positives = 89/171 (52%), Gaps = 16/171 (9%)
Query 3 GHESWNMVINIMVGIRLAGARAMSEPHRAVEPYDFLMKEKFSVLPKTGMVDKKRRKPSLC 62
GH ++ + N++ GIR+A +R S + + P DF +K + + S
Sbjct 382 GHVNFIIAYNMLTGIRVAVSRC-SGIMKPLTPADFRFTKKLAF----DYHGNELTPSSQY 436
Query 63 AVRFIDYAPMVFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSLSELVSEGKSGSLF 122
A +F DY P VFR LRALFG+D Y+ S+ +L SEL S GKSGS F
Sbjct 437 AFKFKDYCPEVFRELRALFGLDPADYLVSLTSKYIL-----------SELNSPGKSGSFF 485
Query 123 YYTTDGRFMIKTVSKETAFFMRSILFDYYKHVSTCTNTMLTRLCGLHALRL 173
YY+ D +++IKT+ +R + +YY HV NT++ + GLH +++
Sbjct 486 YYSRDYKYIIKTIHHSEHIHLRKHIQEYYNHVRDNPNTLICQFYGLHRVKM 536
> At3g56960
Length=779
Score = 92.8 bits (229), Expect = 3e-19, Method: Compositional matrix adjust.
Identities = 66/178 (37%), Positives = 93/178 (52%), Gaps = 26/178 (14%)
Query 3 GHESWNMVINIMVGIRLA-GARA----MSEPHRAVEPYDFLMKEK-FSVLPKTGMVDKKR 56
GH ++ +++N+ +GIR A G +A + H A +P KEK ++ P G K
Sbjct 386 GHRNYELMLNLQLGIRHAVGKQAPVVSLDLKHSAFDP-----KEKVWTRFPPEGT---KY 437
Query 57 RKPSLCA-VRFIDYAPMVFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSLSELVSE 115
P + ++ DY P+VFR LR LF +D Y+ S I GN +L EL S
Sbjct 438 TPPHQSSEFKWKDYCPLVFRSLRKLFKVDPADYMLS----------ICGN-DALRELSSP 486
Query 116 GKSGSLFYYTTDGRFMIKTVSKETAFFMRSILFDYYKHVSTCTNTMLTRLCGLHALRL 173
GKSGS FY T D R+MIKT+ K + +L YY HV NT++ R GLH ++L
Sbjct 487 GKSGSFFYLTNDDRYMIKTMKKSETKVLLRMLAAYYNHVRAFENTLVIRFYGLHCVKL 544
> At1g60890
Length=769
Score = 89.0 bits (219), Expect = 4e-18, Method: Compositional matrix adjust.
Identities = 56/172 (32%), Positives = 89/172 (51%), Gaps = 20/172 (11%)
Query 7 WN--MVINIMVGIRLAGARAMSEPHRAVEPYDFLMKEKFSVL-PKTGMVDKKRRKPSLCA 63
WN +++N+ +GIR + P R V DF + + + P+ G + P +
Sbjct 348 WNHYLMLNLQLGIRYTVGKITPVPPREVRASDFSERARIMMFFPRNG----SQYTPPHKS 403
Query 64 VRFI--DYAPMVFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSLSELVSEGKSGSL 121
+ F DY PMVFR LR +F +D+ Y+ S+ D L E+ S GKSGS+
Sbjct 404 IDFDWKDYCPMVFRNLREMFKLDAADYMMSICGDD-----------GLREISSPGKSGSI 452
Query 122 FYYTTDGRFMIKTVSKETAFFMRSILFDYYKHVSTCTNTMLTRLCGLHALRL 173
FY + D RF+IKT+ + + +L YY+HV NT++T+ G+H ++L
Sbjct 453 FYLSHDDRFVIKTLKRSELKVLLRMLPRYYEHVGDYENTLITKFFGVHRIKL 504
> At1g10900
Length=859
Score = 89.0 bits (219), Expect = 4e-18, Method: Compositional matrix adjust.
Identities = 57/174 (32%), Positives = 88/174 (50%), Gaps = 18/174 (10%)
Query 3 GHESWNMVINIMVGIRLAGARAMSEPHRAVEPYDFLMKEKFSVL-PKTGMVDKKRRKPSL 61
G ++ +++N+ +GIR + P R V DF + + P+ G P
Sbjct 434 GEHNYYLMLNLQLGIRYTVGKITPVPRREVRASDFGKNARTKMFFPRDG----SNFTPPH 489
Query 62 CAVRFI--DYAPMVFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSLSELVSEGKSG 119
+V F DY PMVFR LR +F +D+ Y+ S+ D L+E+ S GKSG
Sbjct 490 KSVDFSWKDYCPMVFRNLRQMFKLDAAEYMMSICGDD-----------GLTEISSPGKSG 538
Query 120 SLFYYTTDGRFMIKTVSKETAFFMRSILFDYYKHVSTCTNTMLTRLCGLHALRL 173
S+FY + D RF+IKT+ K + +L YY+HV NT++T+ G+H + L
Sbjct 539 SIFYLSHDDRFVIKTLKKSELQVLLRMLPKYYEHVGDHENTLITKFFGVHRITL 592
> Hs4505817
Length=540
Score = 74.3 bits (181), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 50/160 (31%), Positives = 77/160 (48%), Gaps = 15/160 (9%)
Query 13 IMVGIRLAGARAMSEPHRAVEPYDFLMKEKFSVLPKTGMVDKKRRKPSLCAVRFIDYAPM 72
I +GI S+P R V DF + E + + + P RF YAP+
Sbjct 33 IQLGIGYTVGNLTSKPERDVLMQDFYVVESVFLPSEGSNLTPAHHYPDF---RFKTYAPL 89
Query 73 VFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSLSELVSEGKSGSLFYYTTDGRFMI 132
FR R LFGI Y+ S+ + L+ EL + G SGSLF+ T+D F+I
Sbjct 90 AFRYFRELFGIKPDDYLYSICSEPLI------------ELSNPGASGSLFFVTSDDEFII 137
Query 133 KTVSKETAFFMRSILFDYYKHVSTCTNTMLTRLCGLHALR 172
KTV + A F++ +L YY +++ T+L + GL+ ++
Sbjct 138 KTVQHKEAEFLQKLLPGYYMNLNQNPRTLLPKFYGLYCMQ 177
> Hs16193739
Length=668
Score = 72.0 bits (175), Expect = 6e-13, Method: Composition-based stats.
Identities = 53/162 (32%), Positives = 77/162 (47%), Gaps = 19/162 (11%)
Query 13 IMVGIRLAGARAMSEPHRAVEPYDFLMKEKFSVLPKTGMVDKKRRKPS--LCAVRFIDYA 70
I +GI S+P R V DF + E P G P+ RF YA
Sbjct 83 IQLGIGYTVGHLSSKPERDVLMQDFYVVESI-FFPSEG----SNLTPAHHFQDFRFKTYA 137
Query 71 PMVFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSLSELVSEGKSGSLFYYTTDGRF 130
P+ FR R LFGI Y+ S+ + L+ EL + G SGSLFY T+D F
Sbjct 138 PVAFRYFRELFGIRPDDYLYSLCNEPLI------------ELSNPGASGSLFYVTSDDEF 185
Query 131 MIKTVSKETAFFMRSILFDYYKHVSTCTNTMLTRLCGLHALR 172
+IKTV + A F++ +L YY +++ T+L + GL+ ++
Sbjct 186 IIKTVMHKEAEFLQKLLPGYYMNLNQNPRTLLPKFYGLYCVQ 227
> 7291493
Length=515
Score = 72.0 bits (175), Expect = 6e-13, Method: Compositional matrix adjust.
Identities = 50/168 (29%), Positives = 81/168 (48%), Gaps = 15/168 (8%)
Query 5 ESWNMVINIMVGIRLAGARAMSEPHRAVEPYDFLMKEKFSVLPKTGMVDKKRRKPSLCAV 64
++ ++ +I +GI+ S+P R + DF E + P+ +
Sbjct 76 QTSQIMGSIQLGIQHTVGSLASKPKRDLLMMDFWEIESITFPPEGSSLTPAHH---YSEF 132
Query 65 RFIDYAPMVFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSLSELVSEGKSGSLFYY 124
R+ YAP+ FR R LFGI PD + ++ S L EL + G SGS+FY
Sbjct 133 RYKIYAPIAFRYFRDLFGIQ---------PDDFMMSMCT---SPLRELSNPGASGSIFYL 180
Query 125 TTDGRFMIKTVSKETAFFMRSILFDYYKHVSTCTNTMLTRLCGLHALR 172
TTD F+IKTV + F++ +L YY +++ T+L + GL+ L+
Sbjct 181 TTDDEFIIKTVQHKEGEFLQKLLPGYYMNLNQNPRTLLPKFFGLYCLQ 228
> Hs4505815
Length=549
Score = 70.1 bits (170), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 40/108 (37%), Positives = 60/108 (55%), Gaps = 12/108 (11%)
Query 65 RFIDYAPMVFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSLSELVSEGKSGSLFYY 124
RF YAP+ FR R LFGI Y+ S+ + L+ EL S G SGSLFY
Sbjct 125 RFKTYAPVAFRYFRELFGIRPDDYLYSLCSEPLI------------ELCSSGASGSLFYV 172
Query 125 TTDGRFMIKTVSKETAFFMRSILFDYYKHVSTCTNTMLTRLCGLHALR 172
++D F+IKTV + A F++ +L YY +++ T+L + GL+ ++
Sbjct 173 SSDDEFIIKTVQHKEAEFLQKLLPGYYMNLNQNPRTLLPKFYGLYCVQ 220
> Hs20550114
Length=512
Score = 69.7 bits (169), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 52/162 (32%), Positives = 77/162 (47%), Gaps = 19/162 (11%)
Query 13 IMVGIRLAGARAMSEPHRAVEPYDFLMKEKFSVLPKTGMVDKKRRKPS--LCAVRFIDYA 70
I +GI ++P R V DF + E P G P+ A RF YA
Sbjct 76 IQLGITYTVGSLSTKPERDVLMQDFYVVESI-FFPSEG----SNLTPAHHYNAFRFKTYA 130
Query 71 PMVFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSLSELVSEGKSGSLFYYTTDGRF 130
P+ FR R LFGI Y+ S+ + L+ EL S G SGSLFY ++D
Sbjct 131 PVAFRYFRELFGIPPDDYLCSLCSEPLI------------ELCSSGASGSLFYVSSDDEL 178
Query 131 MIKTVSKETAFFMRSILFDYYKHVSTCTNTMLTRLCGLHALR 172
+IKT+ + A F++ +L YY ++S T+L + GL+ ++
Sbjct 179 IIKTLQHKEAEFLQKLLPGYYLNLSQNPRTLLPKFFGLYCVQ 220
> CE11121
Length=619
Score = 67.4 bits (163), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 51/158 (32%), Positives = 75/158 (47%), Gaps = 19/158 (12%)
Query 13 IMVGIRLAGARAMSEPHRAVEPYDFLMKEKFSVL--PKTGMVDKKRRKPSLCAVRFIDYA 70
I +GI + S P+R V DF EK ++ P G S RF YA
Sbjct 92 IQLGISNSIGSLASLPNRDVLLQDF---EKVDIVAFPAAGSTITPSH--SFGDFRFRTYA 146
Query 71 PMVFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSLSELVSEGKSGSLFYYTTDGRF 130
P+ FR R LF I ++RS+ + L EL + G SGS+FY + D +F
Sbjct 147 PIAFRYFRNLFHIKPADFLRSICTE------------PLKELSNAGASGSIFYVSQDDQF 194
Query 131 MIKTVSKETAFFMRSILFDYYKHVSTCTNTMLTRLCGL 168
+IKTV + A F++ +L YY +++ T+L + GL
Sbjct 195 IIKTVQHKEADFLQKLLPGYYMNLNQNPRTLLPKFFGL 232
> Hs14745097
Length=435
Score = 66.6 bits (161), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 39/110 (35%), Positives = 60/110 (54%), Gaps = 12/110 (10%)
Query 63 AVRFIDYAPMVFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSLSELVSEGKSGSLF 122
A RF YAP+ FR LFGI Y+ S+ + L+ EL S G SGSLF
Sbjct 83 AFRFKTYAPVAFRYFWELFGIRPDDYLYSLCSEPLI------------ELCSSGASGSLF 130
Query 123 YYTTDGRFMIKTVSKETAFFMRSILFDYYKHVSTCTNTMLTRLCGLHALR 172
Y ++D F++KTV + A F++ +L YY +++ T+L + GL+ ++
Sbjct 131 YVSSDDEFIVKTVRHKEAEFLQKLLPGYYINLNQNPRTLLPKFYGLYCVQ 180
> Hs4505819
Length=416
Score = 65.1 bits (157), Expect = 8e-11, Method: Compositional matrix adjust.
Identities = 47/136 (34%), Positives = 67/136 (49%), Gaps = 14/136 (10%)
Query 38 LMKEKFSVLPKTGMVDKKRRKPSLCA-VRFIDYAPMVFRRLRALFGIDSLCYIRSVGPDQ 96
LM + F K + + K +L + +F +Y PMVFR LR FGID Y SV
Sbjct 66 LMPDDFKAYSKIKVDNHLFNKENLPSRFKFKEYCPMVFRNLRERFGIDDQDYQNSV---- 121
Query 97 LLGNLILGNLSSLSELVSEGKSGSLFYYTTDGRFMIKTVSKETAFFMRSILFDYYKHVST 156
S+ S+G+ G+ F T D RF+IKTVS E M +IL Y++ +
Sbjct 122 --------TRSAPINSDSQGRCGTRFLTTYDRRFVIKTVSSEDVAEMHNILKKYHQFIVE 173
Query 157 C-TNTMLTRLCGLHAL 171
C NT+L + G++ L
Sbjct 174 CHGNTLLPQFLGMYRL 189
> Hs20336256
Length=281
Score = 64.7 bits (156), Expect = 8e-11, Method: Compositional matrix adjust.
Identities = 46/136 (33%), Positives = 67/136 (49%), Gaps = 14/136 (10%)
Query 38 LMKEKFSVLPKTGMVDKKRRKPSLCA-VRFIDYAPMVFRRLRALFGIDSLCYIRSVGPDQ 96
LM + F K + + K +L + +F +Y PMVFR LR FGID Y SV
Sbjct 66 LMPDDFKAYSKIKVDNHLFNKENLPSRFKFKEYCPMVFRNLRERFGIDDQDYQNSVTRSA 125
Query 97 LLGNLILGNLSSLSELVSEGKSGSLFYYTTDGRFMIKTVSKETAFFMRSILFDYYKHVST 156
+ + S+G+ G+ F T D RF+IKTVS E M +IL Y++ +
Sbjct 126 PINS------------DSQGRCGTRFLTTYDRRFVIKTVSSEDVAEMHNILKKYHQFIVE 173
Query 157 CT-NTMLTRLCGLHAL 171
C NT+L + G++ L
Sbjct 174 CHGNTLLPQFLGMYRL 189
> Hs6857820
Length=406
Score = 60.8 bits (146), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 35/110 (31%), Positives = 58/110 (52%), Gaps = 13/110 (11%)
Query 65 RFIDYAPMVFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSLSELVSEGKSGSLFYY 124
+F +Y PMVFR LR FGID + S+ L N S+ +SG+ F+
Sbjct 89 KFKEYCPMVFRNLRERFGIDDQDFQNSLTRSAPLPN------------DSQARSGARFHT 136
Query 125 TTDGRFMIKTVSKETAFFMRSILFDYYKHVSTCTN-TMLTRLCGLHALRL 173
+ D R++IKT++ E M +IL Y++++ C T+L + G++ L +
Sbjct 137 SYDKRYIIKTITSEDVAEMHNILKKYHQYIVECHGITLLPQFLGMYRLNV 186
> 7291195
Length=770
Score = 60.1 bits (144), Expect = 2e-09, Method: Composition-based stats.
Identities = 44/130 (33%), Positives = 59/130 (45%), Gaps = 15/130 (11%)
Query 5 ESWNMVINIMVGIRLAGARAMSEPHRAVEPYDFLMKEKFSVLPKTGMVDKKRRKPSLCAV 64
+S ++ +I +GI+ S+P R + DF E S P +
Sbjct 155 QSKQIMGSIQLGIQHTVGSLASKPKRDLLMNDFWEMETISFPPDGSSITPAHHYNDF--- 211
Query 65 RFIDYAPMVFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSLSELVSEGKSGSLFYY 124
RF YAP+ FR R LFGI PD L ++ S L EL + G SGS+FY
Sbjct 212 RFKVYAPIAFRYFRDLFGI---------APDDFLMSMCA---SPLRELSNPGASGSIFYL 259
Query 125 TTDGRFMIKT 134
TTD F+IKT
Sbjct 260 TTDDEFIIKT 269
> At4g01190
Length=401
Score = 56.2 bits (134), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 32/113 (28%), Positives = 59/113 (52%), Gaps = 12/113 (10%)
Query 60 SLCAVRFIDYAPMVFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSLSELVSEGKSG 119
S+ + DY P+ F ++ L GID Y+ S+ D+ L + +S GK G
Sbjct 25 SITEFDWKDYCPVGFGLIQELEGIDHDDYLLSICTDETL------------KKISSGKIG 72
Query 120 SLFYYTTDGRFMIKTVSKETAFFMRSILFDYYKHVSTCTNTMLTRLCGLHALR 172
++F+ + D RF+IK + K +L YY+H++ +++ TR+ G H+++
Sbjct 73 NVFHISNDNRFLIKILRKSEIKVTLEMLPRYYRHINYHRSSLFTRIFGAHSVK 125
> CE27279
Length=401
Score = 55.5 bits (132), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 42/146 (28%), Positives = 70/146 (47%), Gaps = 15/146 (10%)
Query 32 VEPYDFLMKEKFSVLPKTGMVDKKRRK---PSLCAVRFIDYAPMVFRRLRALFGIDSLCY 88
V P LM + F K + + K PS V+ +Y P VFR LR FG+D+ Y
Sbjct 49 VPPPGLLMPDDFKAYSKVKIDNHNFNKDIMPSHYKVK--EYCPNVFRNLREQFGVDNFEY 106
Query 89 IRSVGPDQLLGNLILGNLSSLSELVSEGKSGSLFYYTTDGRFMIKTVSKETAFFMRSILF 148
+RS+ + +L+ G S S F+ + D +F+IK++ E + S+L
Sbjct 107 LRSLTSYEPEPDLLDG---------SAKDSTPRFFISYDKKFVIKSMDSEAVAELHSVLR 157
Query 149 DYYKH-VSTCTNTMLTRLCGLHALRL 173
+Y+++ V T+L + GL+ L +
Sbjct 158 NYHQYVVEKQGKTLLPQYLGLYRLTI 183
> 7304346
Length=313
Score = 53.1 bits (126), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 36/118 (30%), Positives = 59/118 (50%), Gaps = 15/118 (12%)
Query 55 KRRKPSLCAVRFIDYAPMVFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSLSELVS 114
K PS V+ +Y P+VFR LR FG+D + Y S+ Q + ++ S
Sbjct 79 KENMPSHFKVK--EYCPLVFRNLRERFGVDDVDYRESLTRSQPI------------QIDS 124
Query 115 EGKSGSLFYYTTDGRFMIKTVSKETAFFMRSILFDYYKH-VSTCTNTMLTRLCGLHAL 171
GKSG+ FY + D F+IK+++ E M + L Y+ + V T+L + G++ +
Sbjct 125 SGKSGAQFYQSYDKFFIIKSLTSEEIERMHAFLKQYHPYVVERHGKTLLPQYLGMYRI 182
> Hs21322230
Length=421
Score = 50.1 bits (118), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 45/158 (28%), Positives = 69/158 (43%), Gaps = 17/158 (10%)
Query 14 MVGIRLAGARAMSEPHRAVEPYDFLMKEKFSVLPKTGMVDKKRRKPSLCA-VRFIDYAPM 72
+VG+ L G V P L+ + F K + + + +L + +F +Y P
Sbjct 47 LVGVFLWGVAHSINELSQVPPPVMLLPDDFKASSKIKVNNHLFHRENLPSHFKFKEYCPQ 106
Query 73 VFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSLSELVSEGKSGSLFYYTTDGRFMI 132
VFR LR FGID Y+ S L+ SEG G F + D +I
Sbjct 107 VFRNLRDRFGIDDQDYLVS--------------LTRNPPSESEGSDGR-FLISYDRTLVI 151
Query 133 KTVSKETAFFMRSILFDYYKHVSTC-TNTMLTRLCGLH 169
K VS E M S L +Y++++ C NT+L + G++
Sbjct 152 KEVSSEDIADMHSNLSNYHQYIVKCHGNTLLPQFLGMY 189
> At1g01460
Length=425
Score = 49.7 bits (117), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 29/113 (25%), Positives = 57/113 (50%), Gaps = 12/113 (10%)
Query 60 SLCAVRFIDYAPMVFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSLSELVSEGKSG 119
++ + DY P+ FR ++ L I+ Y++S+ D+ L L S K G
Sbjct 33 TITEFEWKDYCPLGFRLIQELEDINHDEYMKSICNDETLRKL------------STSKVG 80
Query 120 SLFYYTTDGRFMIKTVSKETAFFMRSILFDYYKHVSTCTNTMLTRLCGLHALR 172
++F + D RF+IK + K + +L Y++H+ +T+L++ G H+++
Sbjct 81 NMFLLSKDDRFLIKILRKSEIKVILEMLPGYFRHIHKYRSTLLSKNYGAHSVK 133
> Hs22045987
Length=255
Score = 46.6 bits (109), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 36/123 (29%), Positives = 57/123 (46%), Gaps = 12/123 (9%)
Query 50 GMVDKKRRKPSLCAVRFIDYAPMVFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSL 109
G +D + L F Y P+ F + LFGI C PD L +L L
Sbjct 85 GSLDPTPERHVLIEFPFKTYTPVAFCYFQELFGI---C------PDDDLYSLCS---EPL 132
Query 110 SELVSEGKSGSLFYYTTDGRFMIKTVSKETAFFMRSILFDYYKHVSTCTNTMLTRLCGLH 169
EL S G GSL + + F+IKTV + A F++ ++ Y+ ++ + T+L + GL
Sbjct 133 IELSSSGADGSLLHVSIHNEFIIKTVQHKQAEFLQKLIPGYHIDLNQNSWTLLPKFYGLC 192
Query 170 ALR 172
++
Sbjct 193 CVK 195
> ECU03g0330
Length=296
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 29/103 (28%), Positives = 48/103 (46%), Gaps = 15/103 (14%)
Query 69 YAPMVFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSLSELVSEGKSGSLFYYTTDG 128
Y FR +RAL GI L +G +L I GKSGS ++T D
Sbjct 43 YNHKEFREIRALSGIGGL---HRLGKQYILNEQI------------HGKSGSFLFFTRDF 87
Query 129 RFMIKTVSKETAFFMRSILFDYYKHVSTCTNTMLTRLCGLHAL 171
+F++KT+ K +R ++ +Y +V + L ++ G ++L
Sbjct 88 KFIVKTIRKNEFRCIRGMINEYKVYVLENPMSFLCKILGCYSL 130
> CE18979
Length=1375
Score = 39.3 bits (90), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 23/59 (38%), Positives = 35/59 (59%), Gaps = 5/59 (8%)
Query 116 GKSGSLFYYTTDGRFMIKTVSK-ETAFFMR--SILFDYYKHVSTCTNTMLTRLCGLHAL 171
GKSGS FY T D RF++K +S+ E F++ FDY ++ T + LT LC ++ +
Sbjct 1141 GKSGSFFYRTQDDRFVVKQMSRFEIQSFVKFAPNYFDYL--TTSATESKLTTLCKVYGV 1197
> At1g34260
Length=1456
Score = 32.0 bits (71), Expect = 0.60, Method: Compositional matrix adjust.
Identities = 17/60 (28%), Positives = 30/60 (50%), Gaps = 4/60 (6%)
Query 116 GKSGSLFYYTTDGRFMIKTVSKETAFFMRSILFDYYKHVSTC----TNTMLTRLCGLHAL 171
GKS S+F T D RF++K + K + +Y+K++ T L ++ G+H +
Sbjct 1221 GKSKSVFAKTLDDRFIVKEIKKTEYESFVTFATEYFKYMKDSYDLGNQTCLAKVLGIHQV 1280
> At3g14270
Length=1791
Score = 31.2 bits (69), Expect = 0.95, Method: Compositional matrix adjust.
Identities = 32/104 (30%), Positives = 47/104 (45%), Gaps = 20/104 (19%)
Query 75 RRLRALFGI---DSLCYIRSVGPDQLLGNLILGNLSSLSELVSEGKSGSLFYYTTDGRFM 131
+R AL GI L YIRS+ + G GKS F T D RF+
Sbjct 1505 KRFEALRGICLPSELEYIRSLSRCKKWG-------------AQGGKSNVFFAKTLDDRFI 1551
Query 132 IKTVSK-ETAFFMR--SILFDY-YKHVSTCTNTMLTRLCGLHAL 171
IK V+K E F++ F Y + +ST + T L ++ G++ +
Sbjct 1552 IKQVTKTELESFIKFAPAYFKYLSESISTKSPTCLAKILGIYQV 1595
> ECU01g0440
Length=134
Score = 31.2 bits (69), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 16/34 (47%), Positives = 19/34 (55%), Gaps = 0/34 (0%)
Query 135 VSKETAFFMRSILFDYYKHVSTCTNTMLTRLCGL 168
VS+ TA M+S D YK V T L R+CGL
Sbjct 21 VSQSTAETMKSKAQDSYKRVVEHGKTCLERMCGL 54
> 7302711
Length=1809
Score = 31.2 bits (69), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 16/57 (28%), Positives = 28/57 (49%), Gaps = 4/57 (7%)
Query 116 GKSGSLFYYTTDGRFMIKTVSKETAFFMRSILFDYYKHVSTCTN----TMLTRLCGL 168
GKSGS F T D RF++K ++ Y++++ C T+L ++ G+
Sbjct 1576 GKSGSRFCKTLDDRFVLKEMNSRDMTIFEPFAPKYFEYIDRCQQQQQPTLLAKIFGV 1632
> At4g20090
Length=660
Score = 29.6 bits (65), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 19/71 (26%), Positives = 35/71 (49%), Gaps = 8/71 (11%)
Query 5 ESWNMVINIMVGIRLAGARAMSEPHRAVEPYDFLMKEKFSVLPKTGMVDKKRRKPSLCAV 64
+S+N V+N+++ L HR +E YD+++ ++ + +LC +
Sbjct 149 KSFNSVLNVIINEGLY--------HRGLEFYDYVVNSNMNMNISPNGLSFNLVIKALCKL 200
Query 65 RFIDYAPMVFR 75
RF+D A VFR
Sbjct 201 RFVDRAIEVFR 211
> 7291468
Length=2424
Score = 29.3 bits (64), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 23/83 (27%), Positives = 36/83 (43%), Gaps = 9/83 (10%)
Query 73 VFRRLRA-LFGIDSLCYIRSVGPDQLLGNLILGNLSSLSELVSEGKSGSLFYYTTDGRFM 131
V RR A +F D+L D L ILG L + +S G + + G +
Sbjct 295 VQRRAEASMFTADAL--------DDLNAVTILGGLQKVDSGMSVASLGKMKVSSIGGTIV 346
Query 132 IKTVSKETAFFMRSILFDYYKHV 154
V+ E F+MR L+ +K++
Sbjct 347 DTNVANENKFYMRECLYSKFKYI 369
> At3g48900
Length=337
Score = 29.3 bits (64), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 18/58 (31%), Positives = 26/58 (44%), Gaps = 0/58 (0%)
Query 27 EPHRAVEPYDFLMKEKFSVLPKTGMVDKKRRKPSLCAVRFIDYAPMVFRRLRALFGID 84
EP + P D L ++ V MV+ + S CA + Y F RLRAL ++
Sbjct 11 EPCKKTFPLDHLQNKRVCVDLSCWMVELHKVNKSYCATKEKVYLRGFFHRLRALIALN 68
> Hs14729948
Length=558
Score = 29.3 bits (64), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 17/61 (27%), Positives = 32/61 (52%), Gaps = 6/61 (9%)
Query 116 GKSGSLFYYTTDGRFMIKTVSK-ETAFFMRSILFDYYKHVSTCTN----TMLTRLCGLHA 170
GKSG+ FY T D RF++K + + E F+ Y+ +++ T L ++ G++
Sbjct 320 GKSGAAFYATEDDRFILKQMPRLEVQSFL-DFAPHYFNYITNAVQQKRPTALAKILGVYR 378
Query 171 L 171
+
Sbjct 379 I 379
> At5g07270
Length=519
Score = 29.3 bits (64), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 12/35 (34%), Positives = 20/35 (57%), Gaps = 0/35 (0%)
Query 36 DFLMKEKFSVLPKTGMVDKKRRKPSLCAVRFIDYA 70
DFL +K + LP+TG+V K + +F++ A
Sbjct 134 DFLPSDKLNSLPETGVVTAKNKSEQSALSKFVNKA 168
> At3g17420
Length=447
Score = 29.3 bits (64), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 13/34 (38%), Positives = 20/34 (58%), Gaps = 4/34 (11%)
Query 1 HFGHESWNMVINIMVGIRLAGARAMSEPHRAVEP 34
H GH +W I ++VG A+A++ H A+EP
Sbjct 245 HKGHLTWEARIKVLVGT----AKALAYLHEAIEP 274
> At5g52100
Length=298
Score = 28.9 bits (63), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 17/65 (26%), Positives = 34/65 (52%), Gaps = 2/65 (3%)
Query 102 ILGNLSSLSELVSEGKSGSLFYYTTDGRFMIKTVSKETAFFMRSILFDYYKHVSTCTNTM 161
++ +L+ + +S+GK + TD + + V + TAF M+S++ Y + T +
Sbjct 88 VVSDLTMVLGSISQGKEVGVVIDFTDPSTVYENVKQATAFGMKSVV--YVPRIKPETVSA 145
Query 162 LTRLC 166
L+ LC
Sbjct 146 LSALC 150
Lambda K H
0.327 0.140 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2598880752
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40