bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0690_orf1
Length=122
Score E
Sequences producing significant alignments: (Bits) Value
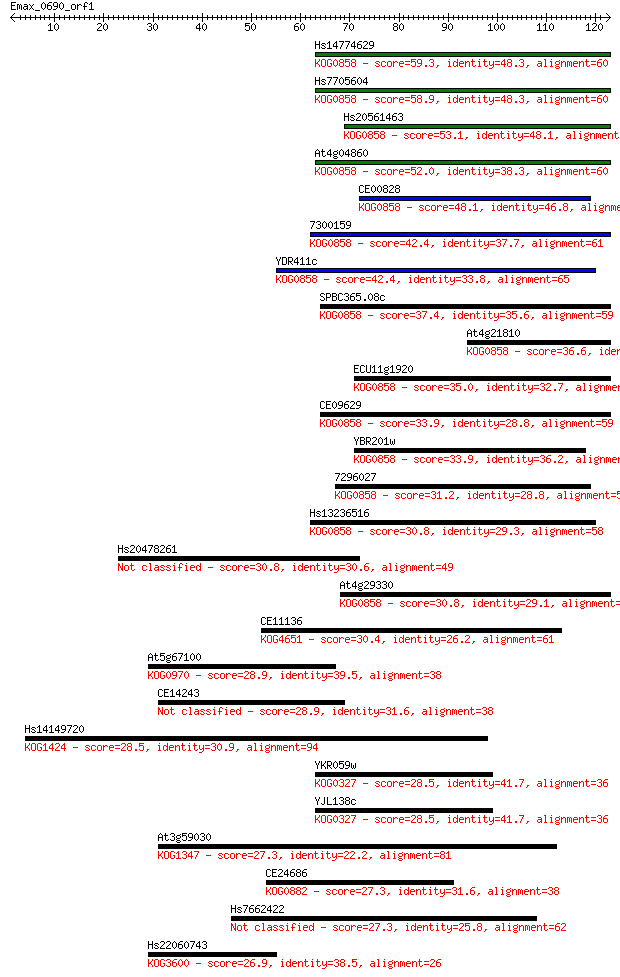
Hs14774629 59.3 2e-09
Hs7705604 58.9 2e-09
Hs20561463 53.1 1e-07
At4g04860 52.0 2e-07
CE00828 48.1 4e-06
7300159 42.4 2e-04
YDR411c 42.4 2e-04
SPBC365.08c 37.4 0.007
At4g21810 36.6 0.011
ECU11g1920 35.0 0.036
CE09629 33.9 0.073
YBR201w 33.9 0.085
7296027 31.2 0.51
Hs13236516 30.8 0.56
Hs20478261 30.8 0.58
At4g29330 30.8 0.71
CE11136 30.4 0.84
At5g67100 28.9 2.4
CE14243 28.9 2.4
Hs14149720 28.5 3.0
YKR059w 28.5 3.2
YJL138c 28.5 3.2
At3g59030 27.3 7.4
CE24686 27.3 7.8
Hs7662422 27.3 8.0
Hs22060743 26.9 8.8
> Hs14774629
Length=239
Score = 59.3 bits (142), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 29/60 (48%), Positives = 38/60 (63%), Gaps = 0/60 (0%)
Query 63 EQVDLFFTHIPPVTRVCLVSSTLLMALCTLEIISPFSLYMNWQLVFTHGQIWRLITCFLF 122
+ + L + IPPV+R + L A LE+I+PF LY N +L+F H QIWRLIT FLF
Sbjct 4 QSLRLEYLQIPPVSRAYTTACVLTTAAVQLELITPFQLYFNPELIFKHFQIWRLITNFLF 63
> Hs7705604
Length=209
Score = 58.9 bits (141), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 29/60 (48%), Positives = 38/60 (63%), Gaps = 0/60 (0%)
Query 63 EQVDLFFTHIPPVTRVCLVSSTLLMALCTLEIISPFSLYMNWQLVFTHGQIWRLITCFLF 122
+ + L + IPPV+R + L A LE+I+PF LY N +L+F H QIWRLIT FLF
Sbjct 4 QSLRLEYLQIPPVSRAYTTACVLTTAAVQLELITPFQLYFNPELIFKHFQIWRLITNFLF 63
> Hs20561463
Length=239
Score = 53.1 bits (126), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 26/54 (48%), Positives = 32/54 (59%), Gaps = 0/54 (0%)
Query 69 FTHIPPVTRVCLVSSTLLMALCTLEIISPFSLYMNWQLVFTHGQIWRLITCFLF 122
F +P VTR + L A LE++SPF LY N LVF Q+WRL+T FLF
Sbjct 10 FLQVPAVTRAYTAACVLTTAAVQLELLSPFQLYFNPHLVFRKFQVWRLVTNFLF 63
> At4g04860
Length=244
Score = 52.0 bits (123), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 23/60 (38%), Positives = 39/60 (65%), Gaps = 0/60 (0%)
Query 63 EQVDLFFTHIPPVTRVCLVSSTLLMALCTLEIISPFSLYMNWQLVFTHGQIWRLITCFLF 122
+ V+ ++ +P +TR L ++ + C+L+IISP++LY+N LV Q WRL+T FL+
Sbjct 3 QAVEEWYKQMPIITRSYLTAAVITTVGCSLDIISPYNLYLNPTLVVKQYQYWRLVTNFLY 62
> CE00828
Length=227
Score = 48.1 bits (113), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 22/47 (46%), Positives = 28/47 (59%), Gaps = 0/47 (0%)
Query 72 IPPVTRVCLVSSTLLMALCTLEIISPFSLYMNWQLVFTHGQIWRLIT 118
+PPVTR + LL LE ++PF LY NW+L+ Q WRLIT
Sbjct 1 MPPVTRFYTGACVLLTTAVHLEFVTPFHLYFNWELIIRKYQFWRLIT 47
> 7300159
Length=261
Score = 42.4 bits (98), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 23/61 (37%), Positives = 30/61 (49%), Gaps = 0/61 (0%)
Query 62 MEQVDLFFTHIPPVTRVCLVSSTLLMALCTLEIISPFSLYMNWQLVFTHGQIWRLITCFL 121
M + F+ IP VTR L L+++SP LY N L+ QIWRL T FL
Sbjct 1 MNALRQFYLEIPVVTRAYTTVCVLTTLAVHLDLVSPLQLYFNPTLIVRKFQIWRLATTFL 60
Query 122 F 122
+
Sbjct 61 Y 61
> YDR411c
Length=341
Score = 42.4 bits (98), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 22/65 (33%), Positives = 33/65 (50%), Gaps = 5/65 (7%)
Query 55 QSVHGPKMEQVDLFFTHIPPVTRVCLVSSTLLMALCTLEIISPFSLYMNWQLVFTHGQIW 114
V GPK F+ +IPP+TR + ++ + L +I+P+ W L F QIW
Sbjct 18 NDVMGPKE-----FWLNIPPITRTLFTLAIVMTIVGRLNLINPWYFIYVWNLTFKKVQIW 72
Query 115 RLITC 119
RL+T
Sbjct 73 RLLTS 77
> SPBC365.08c
Length=224
Score = 37.4 bits (85), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 21/61 (34%), Positives = 37/61 (60%), Gaps = 4/61 (6%)
Query 64 QVDLFFTHIPPVTRVCLV--SSTLLMALCTLEIISPFSLYMNWQLVFTHGQIWRLITCFL 121
Q+ + IPPVTR L+ ++T ++ LC +++SP L +++ LV Q +RL T +L
Sbjct 8 QIQELLSRIPPVTRYILLGTAATTILTLC--QLLSPSMLVLHYPLVVRQKQWYRLFTNYL 65
Query 122 F 122
+
Sbjct 66 Y 66
> At4g21810
Length=225
Score = 36.6 bits (83), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 16/29 (55%), Positives = 21/29 (72%), Gaps = 0/29 (0%)
Query 94 IISPFSLYMNWQLVFTHGQIWRLITCFLF 122
IISP++LY+N LV Q WRL+T FL+
Sbjct 15 IISPYNLYLNPTLVVKQYQFWRLVTNFLY 43
> ECU11g1920
Length=348
Score = 35.0 bits (79), Expect = 0.036, Method: Compositional matrix adjust.
Identities = 17/52 (32%), Positives = 30/52 (57%), Gaps = 1/52 (1%)
Query 71 HIPPVTRVCLVSSTLLMALCTLEIISPFSLYMNWQLVFTHGQIWRLITCFLF 122
+PP+TR + + + L ++ +SP+SLY + L +IWR+ T FL+
Sbjct 18 RVPPITRYMTLLISAVALLVYVDAVSPYSLYYS-PLFLKRLEIWRVFTSFLY 68
> CE09629
Length=245
Score = 33.9 bits (76), Expect = 0.073, Method: Compositional matrix adjust.
Identities = 17/59 (28%), Positives = 30/59 (50%), Gaps = 0/59 (0%)
Query 64 QVDLFFTHIPPVTRVCLVSSTLLMALCTLEIISPFSLYMNWQLVFTHGQIWRLITCFLF 122
++ F IP VTR ++ST++ L I+ +++ W LV Q WR +T ++
Sbjct 2 DLENFLLGIPIVTRYWFLASTIIPLLGRFGFINVQWMFLQWDLVVNKFQFWRPLTALIY 60
> YBR201w
Length=211
Score = 33.9 bits (76), Expect = 0.085, Method: Compositional matrix adjust.
Identities = 17/47 (36%), Positives = 26/47 (55%), Gaps = 0/47 (0%)
Query 71 HIPPVTRVCLVSSTLLMALCTLEIISPFSLYMNWQLVFTHGQIWRLI 117
IP VTR+ + +L L +L I+ P + ++ LVF GQ RL+
Sbjct 11 DIPLVTRLWTIGCLVLSGLTSLRIVDPGKVVYSYDLVFKKGQYGRLL 57
> 7296027
Length=245
Score = 31.2 bits (69), Expect = 0.51, Method: Compositional matrix adjust.
Identities = 15/52 (28%), Positives = 30/52 (57%), Gaps = 0/52 (0%)
Query 67 LFFTHIPPVTRVCLVSSTLLMALCTLEIISPFSLYMNWQLVFTHGQIWRLIT 118
+++ +P TR L ++ +L LC ++I L+++ VF+ Q+WR +T
Sbjct 5 VWYRSLPRFTRYWLTATVVLSMLCRFDVIPLHWLHLDRSAVFSKLQLWRCMT 56
> Hs13236516
Length=251
Score = 30.8 bits (68), Expect = 0.56, Method: Compositional matrix adjust.
Identities = 17/58 (29%), Positives = 28/58 (48%), Gaps = 0/58 (0%)
Query 62 MEQVDLFFTHIPPVTRVCLVSSTLLMALCTLEIISPFSLYMNWQLVFTHGQIWRLITC 119
M + +F IP +TR ++ + + L +ISP L++ + QIWR IT
Sbjct 1 MSDIGDWFRSIPAITRYWFAATVAVPLVGKLGLISPAYLFLWPEAFLYRFQIWRPITA 58
> Hs20478261
Length=100
Score = 30.8 bits (68), Expect = 0.58, Method: Compositional matrix adjust.
Identities = 15/49 (30%), Positives = 27/49 (55%), Gaps = 4/49 (8%)
Query 23 RWSAVSPATPEHRESHGHFPVLCFSSVDPFFWQSVHGPKMEQVDLFFTH 71
RW++ + T EHRES +PV CF +P +Q++ ++ + + H
Sbjct 9 RWTSTANGT-EHRES---YPVDCFLDDEPSHYQNLFTGGRQEEESYIEH 53
> At4g29330
Length=281
Score = 30.8 bits (68), Expect = 0.71, Method: Compositional matrix adjust.
Identities = 16/55 (29%), Positives = 26/55 (47%), Gaps = 0/55 (0%)
Query 68 FFTHIPPVTRVCLVSSTLLMALCTLEIISPFSLYMNWQLVFTHGQIWRLITCFLF 122
F+ +PP+T+ L +++P + + +LV QIWRLIT F
Sbjct 22 FYNSLPPITKAYGTLCFFTTVATQLGLVAPVHIALIPELVLKQFQIWRLITNLFF 76
> CE11136
Length=304
Score = 30.4 bits (67), Expect = 0.84, Method: Compositional matrix adjust.
Identities = 16/61 (26%), Positives = 29/61 (47%), Gaps = 8/61 (13%)
Query 52 FFWQSVHGPKMEQVDLFFTHIPPVTRVCLVSSTLLMALCTLEIISPFSLYMNWQLVFTHG 111
F +QS H + D+ +T+ VS TL+ + + F+LY+ WQ + + G
Sbjct 191 FDYQSRHAAITQLTDILYTN--------RVSDTLITKISNESMSKIFTLYLQWQCLVSAG 242
Query 112 Q 112
+
Sbjct 243 E 243
> At5g67100
Length=1492
Score = 28.9 bits (63), Expect = 2.4, Method: Composition-based stats.
Identities = 15/38 (39%), Positives = 17/38 (44%), Gaps = 12/38 (31%)
Query 29 PATPEHRESHGHFPVLCFSSVDPFFWQSVHGPKMEQVD 66
PATPE ES D FW +H PK +Q D
Sbjct 1325 PATPETEES------------DSTFWLKLHCPKCQQED 1350
> CE14243
Length=357
Score = 28.9 bits (63), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 12/38 (31%), Positives = 20/38 (52%), Gaps = 0/38 (0%)
Query 31 TPEHRESHGHFPVLCFSSVDPFFWQSVHGPKMEQVDLF 68
T +H+ + F + S + FFWQ+ + K + VD F
Sbjct 66 TSQHKNTKFAFKIKALSFQNKFFWQTRYLEKQDAVDHF 103
> Hs14149720
Length=658
Score = 28.5 bits (62), Expect = 3.0, Method: Composition-based stats.
Identities = 29/108 (26%), Positives = 40/108 (37%), Gaps = 17/108 (15%)
Query 4 ALVIYLFCGNTGQLLCVGGRWSAVSPATPEHRESHGHF------PVLCFSSVDPFFWQSV 57
LV Y G + + + G ATP H + HF P LC S
Sbjct 390 GLVGYPNVGKSSTINTIMGNKKVSVSATPGHTK---HFQTLYVEPGLCLCDCPGLVMPSF 446
Query 58 HGPKMEQ-------VDLFFTHIPPVTRVCL-VSSTLLMALCTLEIISP 97
K E +D H+PPV+ VC + +L A + II+P
Sbjct 447 VSTKAEMTCSGILPIDQMRDHVPPVSLVCQNIPRHVLEATYGINIITP 494
> YKR059w
Length=395
Score = 28.5 bits (62), Expect = 3.2, Method: Composition-based stats.
Identities = 15/36 (41%), Positives = 22/36 (61%), Gaps = 2/36 (5%)
Query 63 EQVDLFFTHIPPVTRVCLVSSTLLMALCTLEIISPF 98
EQ+ FT +PP T+V L+S+T M LE+ + F
Sbjct 182 EQIYQIFTLLPPTTQVVLLSAT--MPNDVLEVTTKF 215
> YJL138c
Length=395
Score = 28.5 bits (62), Expect = 3.2, Method: Composition-based stats.
Identities = 15/36 (41%), Positives = 22/36 (61%), Gaps = 2/36 (5%)
Query 63 EQVDLFFTHIPPVTRVCLVSSTLLMALCTLEIISPF 98
EQ+ FT +PP T+V L+S+T M LE+ + F
Sbjct 182 EQIYQIFTLLPPTTQVVLLSAT--MPNDVLEVTTKF 215
> At3g59030
Length=507
Score = 27.3 bits (59), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 18/81 (22%), Positives = 36/81 (44%), Gaps = 5/81 (6%)
Query 31 TPEHRESHGHFPVLCFSSVDPFFWQSVHGPKMEQVDLFFTHIPPVTRVCLVSSTLLMALC 90
+P +E+ F F + P+F +V M +++++ + ++S L
Sbjct 255 SPNCKETWTGFSTRAFRGIWPYFKLTVASAVMLCLEIWYNQ-----GLVIISGLLSNPTI 309
Query 91 TLEIISPFSLYMNWQLVFTHG 111
+L+ IS Y+NW + F G
Sbjct 310 SLDAISICMYYLNWDMQFMLG 330
> CE24686
Length=629
Score = 27.3 bits (59), Expect = 7.8, Method: Composition-based stats.
Identities = 12/39 (30%), Positives = 19/39 (48%), Gaps = 1/39 (2%)
Query 53 FWQSVHGPKMEQVDLFFTHIPPVTRVCL-VSSTLLMALC 90
FW+ H +E V F H+ + +C + TLL +C
Sbjct 79 FWKKKHSEGVEFVKHFRCHLSEFSHICANIDGTLLATVC 117
> Hs7662422
Length=648
Score = 27.3 bits (59), Expect = 8.0, Method: Composition-based stats.
Identities = 16/62 (25%), Positives = 30/62 (48%), Gaps = 2/62 (3%)
Query 46 FSSVDPFFWQSVHGPKMEQVDLFFTHIPPVTRVCLVSSTLLMALCTLEIISPFSLYMNWQ 105
+++ PF SV + FF ++ + LVS++ M LC L + P ++ NW
Sbjct 2 LTTLKPFGSVSVESKMNNKAGSFFWNLRQFS--TLVSTSRTMRLCCLGLCKPKIVHSNWN 59
Query 106 LV 107
++
Sbjct 60 IL 61
> Hs22060743
Length=2012
Score = 26.9 bits (58), Expect = 8.8, Method: Compositional matrix adjust.
Identities = 10/26 (38%), Positives = 14/26 (53%), Gaps = 0/26 (0%)
Query 29 PATPEHRESHGHFPVLCFSSVDPFFW 54
P P+ +SH H P + VDPF +
Sbjct 1464 PTEPDSADSHAHPPAVVIYMVDPFTY 1489
Lambda K H
0.332 0.143 0.501
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1194805952
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40