bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0696_orf2
Length=205
Score E
Sequences producing significant alignments: (Bits) Value
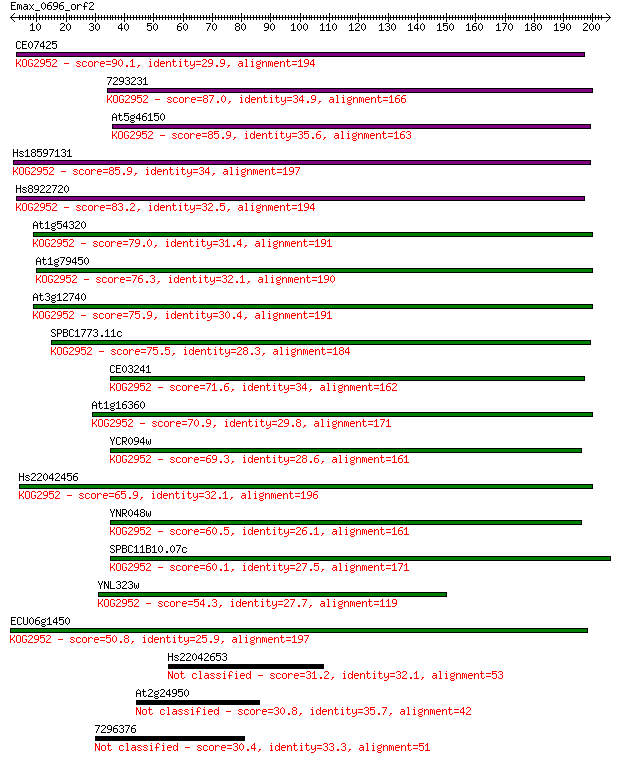
CE07425 90.1 3e-18
7293231 87.0 2e-17
At5g46150 85.9 5e-17
Hs18597131 85.9 5e-17
Hs8922720 83.2 3e-16
At1g54320 79.0 6e-15
At1g79450 76.3 4e-14
At3g12740 75.9 5e-14
SPBC1773.11c 75.5 6e-14
CE03241 71.6 1e-12
At1g16360 70.9 2e-12
YCR094w 69.3 4e-12
Hs22042456 65.9 6e-11
YNR048w 60.5 2e-09
SPBC11B10.07c 60.1 3e-09
YNL323w 54.3 2e-07
ECU06g1450 50.8 2e-06
Hs22042653 31.2 1.3
At2g24950 30.8 1.8
7296376 30.4 2.5
> CE07425
Length=348
Score = 90.1 bits (222), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 58/196 (29%), Positives = 92/196 (46%), Gaps = 19/196 (9%)
Query 3 NSIVECKVNYTDMDGDVSLKIDASTCTDTSITEITGDVLLYYELSNYFQNHRHFMKSRSD 62
+++ E V YT+ L+I+ D GDV LYY L NY+QNHR ++KSR+D
Sbjct 76 DAVSEFTVEYTNCLSPCQLQINLPNAFD-------GDVFLYYNLENYYQNHRRYVKSRND 128
Query 63 KQLEGKVYTAASEVRTACDPRVESSDGRVLHPCGLSAGSVFTDVFSAVEEDEITEIEL-D 121
+Q G + + V+ ++ + + + PCG A S+F D F+ + + +
Sbjct 129 QQYLGDL----TNVKDCAPFDIDPATKKPIAPCGAIANSIFNDTFTLAHRADTGIVTMVP 184
Query 122 ESREAICWDWDL-STFKNPSSEEINAAAPQVDFWLFNQTYASALLMDKPGVGWGVENSHF 180
+ + + W+ D FKNP + N FN T P G EN F
Sbjct 185 VTTQGVIWNVDKDRKFKNPPLNDGNLCD------AFNNTTKPPNWSKNPCEVGGFENVDF 238
Query 181 IVWVRESPLPVFRKVY 196
IVW+R + LP F+K++
Sbjct 239 IVWMRTAALPYFKKLW 254
> 7293231
Length=357
Score = 87.0 bits (214), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 58/168 (34%), Positives = 87/168 (51%), Gaps = 11/168 (6%)
Query 34 TEITGDVLLYYELSNYFQNHRHFMKSRSDKQLEGKVYTAASEVRTACDPRVESSD-GRVL 92
++ G V +YY L+NY+QNHR ++KSR D+QL G + S T C P D G+ +
Sbjct 107 SDFNGVVYMYYGLTNYYQNHRRYVKSRDDEQLLGHLSQTPS---TDCAPFAYDPDSGKPI 163
Query 93 HPCGLSAGSVFTDVFSAVEEDEITEIELDESREAICWDWDLST-FKNPSSEEINAAAPQV 151
PCG A S+F D + ++ +EI+L ++ I W D F+NP +N +
Sbjct 164 APCGAIANSLFNDTLTLLQGG--SEIKLLKT--GIAWPSDKRVKFRNPEG-NLNVSLEGF 218
Query 152 DFWLFNQTYASALLMDKPGVGWGVENSHFIVWVRESPLPVFRKVYGRF 199
+F Q + L + P G +N IVW+R + LP FRK+Y R
Sbjct 219 SKPIFWQKGLADLDPENPDNN-GFQNEDLIVWMRTAALPSFRKLYRRL 265
> At5g46150
Length=329
Score = 85.9 bits (211), Expect = 5e-17, Method: Compositional matrix adjust.
Identities = 58/164 (35%), Positives = 84/164 (51%), Gaps = 21/164 (12%)
Query 36 ITGDVLLYYELSNYFQNHRHFMKSRSDKQ-LEGKVYTAASEVRTACDPRVESSDGRVLHP 94
+ + +YY+L NY+QNHR ++KSRSD+Q L G Y+ S +C+P ESS+G + P
Sbjct 114 MKAPIFIYYQLDNYYQNHRRYVKSRSDQQLLHGLEYSHTS----SCEPE-ESSNGLPIVP 168
Query 95 CGLSAGSVFTDVFSAVEEDEITEIELDESREAICWDWDLSTFKNPSSEEINAAAPQVDFW 154
CGL A S+F D F+ E +L+ SR I W D + ++F
Sbjct 169 CGLIAWSMFNDTFTFSRE----RTKLNVSRNNIAWKSD-------REHKFGKNVYPINFQ 217
Query 155 LFNQTYASALLMDKPGVGWGVENSHFIVWVRESPLPVFRKVYGR 198
N T +D P + + FIVW+R + L FRK+YGR
Sbjct 218 --NGTLIGGAKLD-PKIPLS-DQEDFIVWMRAAALLSFRKLYGR 257
> Hs18597131
Length=351
Score = 85.9 bits (211), Expect = 5e-17, Method: Compositional matrix adjust.
Identities = 67/229 (29%), Positives = 94/229 (41%), Gaps = 49/229 (21%)
Query 2 NNSIVECKVNYTDMDGDVSLKIDASTCTDTSITE-------------ITGDVLLYYELSN 48
+N I E + +YT G + + A+ ++ G V LYYEL+N
Sbjct 57 SNGIKELEYDYTGDPGTGNCSVCAAAGQGRALPPPCSCAWYFSLPELFQGPVYLYYELTN 116
Query 49 YFQNHRHFMKSRSDKQLEGKVYTAASEVRTACDPRVESSDGRVLHPCGLSAGSVFTDVFS 108
++QN+R + SR D QL G + +A C P S+ G + PCG A S+F D FS
Sbjct 117 FYQNNRRYGVSRDDAQLSG-LPSALRHPVNECAPYQRSAAGLPIAPCGAIANSLFNDSFS 175
Query 109 AVEEDE----ITEIELDESREAICWDWDLST-FKNPSSEEINAAAPQVDFWLFNQTYASA 163
+ + E+ LD R I W D F+NP L N + A A
Sbjct 176 LWHQRQPGGPYVEVPLD--RSGIAWWTDYHVKFRNPP--------------LVNGSLALA 219
Query 164 LLMDKPGVGW--------------GVENSHFIVWVRESPLPVFRKVYGR 198
P W G N F+VW+R + LP FRK+Y R
Sbjct 220 FQGTAPPPNWRRPVYELSPDPNNTGFINQDFVVWMRTAALPTFRKLYAR 268
> Hs8922720
Length=361
Score = 83.2 bits (204), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 63/209 (30%), Positives = 96/209 (45%), Gaps = 23/209 (11%)
Query 3 NSIVECKVNYTDMDGD------VSLKIDASTCTDTSITE--ITGDVLLYYELSNYFQNHR 54
N+I E +++YT + +S + CT E G+V +YY LSN++QNHR
Sbjct 73 NNIREIEIDYTGTEPSSPCNKCLSPDVTPCFCTINFTLEKSFEGNVFMYYGLSNFYQNHR 132
Query 55 HFMKSRSDKQLEGKVYTAASEVRTACDPRVESSDGRVLHPCGLSAGSVFTDVFSA-VEED 113
++KSR D QL G +A C+P + D + + PCG A S+F D + +
Sbjct 133 RYVKSRDDSQLNGDS-SALLNPSKECEPYRRNED-KPIAPCGAIANSMFNDTLELFLIGN 190
Query 114 EITEIELDESREAICWDWDLST-FKNPSS-----EEINAAAPQVDFWLFNQTYASALLMD 167
+ I + ++ I W D + F+NP E V+ WL ++D
Sbjct 191 DSYPIPIALKKKGIAWWTDKNVKFRNPPGGDNLEERFKGTTKPVN-WL-----KPVYMLD 244
Query 168 KPGVGWGVENSHFIVWVRESPLPVFRKVY 196
G N FIVW+R + LP FRK+Y
Sbjct 245 SDPDNNGFINEDFIVWMRTAALPTFRKLY 273
> At1g54320
Length=349
Score = 79.0 bits (193), Expect = 6e-15, Method: Compositional matrix adjust.
Identities = 60/193 (31%), Positives = 91/193 (47%), Gaps = 28/193 (14%)
Query 9 KVNYTDMDGDVSLKIDASTCT-DTSITE-ITGDVLLYYELSNYFQNHRHFMKSRSDKQLE 66
KV Y DGD C D +T+ + + +YY+L N++QNHR ++KSRSD QL
Sbjct 95 KVAYIQGDGD-------KVCNRDLKVTKRMKQPIYVYYQLENFYQNHRRYVKSRSDSQLR 147
Query 67 GKVYTAASEVRTACDPRVESSDGRVLHPCGLSAGSVFTDVFSAVEEDEITEIELDESREA 126
Y +AC P + G+ + PCGL A S+F D ++ + + L +++
Sbjct 148 STKYENQI---SACKPE-DDVGGQPIVPCGLIAWSLFNDTYALSRNN----VSLAVNKKG 199
Query 127 ICWDWDLSTFKNPSSEEINAAAPQVDFWLFNQTYASALLMDKPGVGWGVENSHFIVWVRE 186
I W D + N P+ +F N T + L P + E IVW+R
Sbjct 200 IAWKSD------KEHKFGNKVFPK-NFQKGNITGGATL---DPRIPLS-EQEDLIVWMRT 248
Query 187 SPLPVFRKVYGRF 199
+ LP FRK+YG+
Sbjct 249 AALPTFRKLYGKI 261
> At1g79450
Length=350
Score = 76.3 bits (186), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 61/192 (31%), Positives = 88/192 (45%), Gaps = 28/192 (14%)
Query 10 VNYTDMDGDVSLKIDASTCTDTSITEITGDVLLYYELSNYFQNHRHFMKSRSDKQLEGKV 69
V Y +GD KI T T T + V +YY+L N++QNHR ++KSR+D QL
Sbjct 96 VAYIQGEGD---KICKRTITVTK--AMKHPVYVYYQLENFYQNHRRYVKSRNDAQLRSP- 149
Query 70 YTAASEVRTACDPRVESSDGRVLHPCGLSAGSVFTDVFSAVEEDEITEIELDESREAICW 129
+V+T C P ++ G + PCGL A S+F D +S + +L +++ I W
Sbjct 150 -KEEHDVKT-CAPE-DNVGGEPIVPCGLVAWSLFNDTYSFSRNSQ----QLLVNKKGISW 202
Query 130 DWDLST--FKNPSSEEINAAAPQVDFWLFNQTYASALLMDKPGVGWGVENSHFIVWVRES 187
D KN + AP L + KP E IVW+R +
Sbjct 203 KSDRENKFGKNVFPKNFQKGAP---------IGGGTLNISKP----LSEQEDLIVWMRTA 249
Query 188 PLPVFRKVYGRF 199
LP FRK+YG+
Sbjct 250 ALPTFRKLYGKI 261
> At3g12740
Length=350
Score = 75.9 bits (185), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 58/194 (29%), Positives = 89/194 (45%), Gaps = 30/194 (15%)
Query 9 KVNYTDMDGDVSLKIDASTCTDTSIT--EITGDVLLYYELSNYFQNHRHFMKSRSDKQLE 66
KV Y G+ S CT T I + + +YY+L N++QNHR ++KSRSD QL
Sbjct 96 KVAYIQGTGNKS-------CTRTLIVPKRMKQPIYVYYQLENFYQNHRRYVKSRSDSQLR 148
Query 67 GKVYTAASEVRT-ACDPRVESSDGRVLHPCGLSAGSVFTDVFSAVEEDEITEIELDESRE 125
+ E + AC P + G+ + PCGL A S+F D + ++ L +++
Sbjct 149 ----SVKDENQIDACKPE-DDFGGQPIVPCGLIAWSLFNDTYVLSRNNQ----GLTVNKK 199
Query 126 AICWDWDLSTFKNPSSEEINAAAPQVDFWLFNQTYASALLMDKPGVGWGVENSHFIVWVR 185
I W D + +F N T ++L +KP + IVW+R
Sbjct 200 GIAWKSD-------KEHKFGKNVFPKNFQKGNLTGGASLDPNKP----LSDQEDLIVWMR 248
Query 186 ESPLPVFRKVYGRF 199
+ LP FRK+YG+
Sbjct 249 TAALPTFRKLYGKI 262
> SPBC1773.11c
Length=396
Score = 75.5 bits (184), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 52/184 (28%), Positives = 84/184 (45%), Gaps = 7/184 (3%)
Query 15 MDGDVSLKIDASTCTDTSITEITGDVLLYYELSNYFQNHRHFMKSRSDKQLEGKVYTAAS 74
+D D+ + ++ T + + + +YY L+N+FQNHR + KS +KQL+G TA
Sbjct 123 LDKDLDMDVNYCIIRFTVPSVLKAPIFIYYRLTNFFQNHRRYAKSVDEKQLQGVALTADE 182
Query 75 EVRTACDPRVESSDGRVLHPCGLSAGSVFTDVFSAVEEDEITEIELDESREAICWDWDLS 134
C P + D + +PCGL A S+F D FS++ + + S + I W D
Sbjct 183 VKGGNCFPLEVNEDDKPYYPCGLIANSLFNDTFSSLRLLDDNSV-YTFSTKNIAWASDKR 241
Query 135 TFKNPSSEEINAAAPQVDFWLFNQTYASALLMDKPGVGWGVENSHFIVWVRESPLPVFRK 194
F + + A P + Y + + D +EN VW+R + LP F K
Sbjct 242 RFLKTNYSPDDVAPPPNWVLRYPDGYTESNMPDLS----TMENLQ--VWMRTAGLPTFSK 295
Query 195 VYGR 198
+ R
Sbjct 296 LAMR 299
> CE03241
Length=361
Score = 71.6 bits (174), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 55/176 (31%), Positives = 82/176 (46%), Gaps = 21/176 (11%)
Query 35 EITGDVLLYYELSNYFQNHRHFMKSRSDKQLEGKVYTAASEVRTACDP--RVESSDGRV- 91
+ TG+V YY LS ++QN+R + SR+D+QL GKV CDP V+ + +V
Sbjct 107 DYTGEVKFYYGLSKFYQNNRLYFNSRNDQQLRGKVTET-----DGCDPLEYVDVNGTKVP 161
Query 92 LHPCGLSAGSVFTDVFSAV-----EEDEITEIELDESREAICWDWDLSTFKNPSSEE--- 143
+ PCG A S+F D F + +T + +R + F+NP E
Sbjct 162 IAPCGKVADSMFNDTFELFYINDKASNAVTRVPW-TTRGVLGATEMKRKFRNPIRAENQT 220
Query 144 ---INAAAPQVDFWLFNQTYASALLMDKPGVGWGVENSHFIVWVRESPLPVFRKVY 196
+ A W + +D P VG G EN F+VW++ + LP FRK+Y
Sbjct 221 LCDVFAGTMPPPSWRYPICQLGLNSID-PDVGIGFENIDFMVWMKVAALPKFRKLY 275
> At1g16360
Length=316
Score = 70.9 bits (172), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 51/172 (29%), Positives = 80/172 (46%), Gaps = 21/172 (12%)
Query 29 TDTSITEITGDVLLYYELSNYFQNHRHFMKSRSDKQLEGKVYTAASEVRT-ACDPRVESS 87
T T + V +YY+L NY+QNHR ++KSR D QL + E T +C P ++
Sbjct 76 TITVTKTMKNPVYVYYQLENYYQNHRRYVKSRQDGQLR----SPKDEHETKSCAPE-DTL 130
Query 88 DGRVLHPCGLSAGSVFTDVFSAVEEDEITEIELDESREAICWDWDLSTFKNPSSEEINAA 147
G+ + PCGL A S+F D + ++ +L +++ I W K+ +
Sbjct 131 GGQPIVPCGLVAWSLFNDTYDFTRNNQ----KLPVNKKDISW-------KSDRESKFGKN 179
Query 148 APQVDFWLFNQTYASALLMDKPGVGWGVENSHFIVWVRESPLPVFRKVYGRF 199
+F + +L D P E IVW+R + LP FRK+YG+
Sbjct 180 VFPKNFQKGSLIGGKSLDQDIPLS----EQEDLIVWMRTAALPTFRKLYGKI 227
> YCR094w
Length=391
Score = 69.3 bits (168), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 46/168 (27%), Positives = 78/168 (46%), Gaps = 22/168 (13%)
Query 35 EITGDVLLYYELSNYFQNHRHFMKSRSDKQLEGKVYTAASEVRTACDPRVESSDGRVLHP 94
+I + +YY+++N++QNHR +++S KQ+ G+ ++ T+C P + S + ++++P
Sbjct 132 DIKKSIFIYYKITNFYQNHRRYVQSFDTKQILGEP-IKKDDLDTSCSP-IRSREDKIIYP 189
Query 95 CGLSAGSVFTDVFSAVEEDEITEIELDESREAICWDWDLSTFKNPSSEEINAAAPQVDFW 154
CGL A S+F D FS V + + + + I W D FK + P W
Sbjct 190 CGLIANSMFNDTFSQVLSGIDDTEDYNLTNKHISWSIDRHRFKTTKYNASDIVPPPN--W 247
Query 155 LFNQTYASALLMDKPGVGWGVEN-------SHFIVWVRESPLPVFRKV 195
M K G+ EN F VW+R + P F K+
Sbjct 248 -----------MKKYPDGYTDENLPDIHTWEEFQVWMRTAAFPKFYKL 284
> Hs22042456
Length=314
Score = 65.9 bits (159), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 63/218 (28%), Positives = 97/218 (44%), Gaps = 41/218 (18%)
Query 4 SIVECKVNYTDMDGDVS-LKIDAST----CTDTSIT-----EITGDVLLYYELSNYFQNH 53
S E ++NYT + + + L+ +AS CT SI ++ G+V +YY+L ++QN
Sbjct 69 STQEIEINYTRICANCAKLRENASNFDKECT-CSIPFYLSGKMMGNVYMYYKLYGFYQNL 127
Query 54 RHFMKSRSDKQLEGK--------VYTAASEVRTA---CDPRVESSDGRVLHPCGLSAGSV 102
+++SRS++QL GK A S A C P S + + PCG A S+
Sbjct 128 YLYIRSRSNRQLVGKDVKSLPKAFLIAVSSCIHAVEDCAPFKMSDNKTPIVPCGAIANSM 187
Query 103 FTD-VFSAVEEDEITEIELDESREAICWDWDLSTFKNPSSEEINAAAPQVDFWLFNQTYA 161
F D + + + +I++ + + W W T K P N P D
Sbjct 188 FNDTIILSHNINSSVQIKVPMLKSRLTW-WTDKTTKPP-----NWPKPIYD--------- 232
Query 162 SALLMDKPGVGWGVENSHFIVWVRESPLPVFRKVYGRF 199
L K G N FIVW+R + P F+K+YGR
Sbjct 233 ---LDKKDPRNNGFLNDDFIVWMRAAAFPTFKKLYGRL 267
> YNR048w
Length=393
Score = 60.5 bits (145), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 42/162 (25%), Positives = 71/162 (43%), Gaps = 10/162 (6%)
Query 35 EITGDVLLYYELSNYFQNHRHFMKSRSDKQLEGKVYTAASEVRTACDPRVESSDGRVLHP 94
I +YY L+N+ QN+R +++S QL+GK +++ CDP + + + + + P
Sbjct 135 HIKKSTYVYYRLTNFNQNYREYVQSLDLDQLKGKALI-GNDLDPNCDP-LRTVENKTIFP 192
Query 95 CGLSAGSVFTDVFSAVEEDEITEIELDESREAICWDWDLSTFKNPSSEEINAAAPQVDFW 154
CGL A S+F D F + + + I WD D + + P
Sbjct 193 CGLIANSMFNDTFGTTLTGVNDTADYLLTTKGIAWDTDSHRYGKTEYNASDIVPPPNWAK 252
Query 155 LFNQTYASALLMDKPGVGWGVEN-SHFIVWVRESPLPVFRKV 195
LF Y + D ++N F +W+R + LP F K+
Sbjct 253 LFPNGYTDDNIPD-------LQNWEQFKIWMRTAALPNFYKL 287
> SPBC11B10.07c
Length=371
Score = 60.1 bits (144), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 47/177 (26%), Positives = 74/177 (41%), Gaps = 12/177 (6%)
Query 35 EITGDVLLYYELSNYFQNHRHFMKSRSDKQLEGKVYTAAS-EVRTACDPRVESSDGRVLH 93
E+T V +Y L N++QNHR + S QL G+ T A + C P + +G+ +
Sbjct 120 EMTSPVFAFYRLKNFYQNHRRYTVSADMFQLLGEARTVAQLKSYGFCKPLEANEEGKPYY 179
Query 94 PCGLSAGSVFTDVFSAVEE----DEITEIEL-DESREAICWDWDLSTFKNPSSEEINAAA 148
PCG+ A S+F D +S++ D + L + + W D +K
Sbjct 180 PCGIIANSLFNDSYSSLLRYESFDSSNSLGLYNMTTNGTAWPEDRERYKKTKYNASQIVP 239
Query 149 PQVDFWLFNQTYASALLMDKPGVGWGVENSHFIVWVRESPLPVFRKVYGRFVAQPLK 205
P +F Y + D W F +W+R + LP F K+ R V L+
Sbjct 240 PPNWAKMFPNGYTDDNIPDV--STWDA----FQIWMRAAALPTFSKLALRNVTTALQ 290
> YNL323w
Length=414
Score = 54.3 bits (129), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 33/123 (26%), Positives = 56/123 (45%), Gaps = 4/123 (3%)
Query 31 TSITEITGDVLLYYELSNYFQNHRHFMKSRSDKQLEGK---VYTAASEVRTACDPRVESS 87
T+ +++ +V L Y L + NHR ++ S S+ Q+ G+ T C P +++
Sbjct 164 TTPSDMKNNVYLNYVLEKFAANHRRYVLSFSEDQIRGEDASYETVHDATGINCKPLSKNA 223
Query 88 DGRVLHPCGLSAGSVFTDVFS-AVEEDEITEIELDESREAICWDWDLSTFKNPSSEEINA 146
DG++ +PCGL A S+F D F + T + + I W+ D +K
Sbjct 224 DGKIYYPCGLIANSMFNDTFPLQLTNVGDTSNNYSLTNKGINWESDKKRYKKTKYNYTQI 283
Query 147 AAP 149
A P
Sbjct 284 APP 286
> ECU06g1450
Length=270
Score = 50.8 bits (120), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 51/198 (25%), Positives = 84/198 (42%), Gaps = 41/198 (20%)
Query 1 VNNSIVECKVNYTD-MDGDVSLKIDASTCTDTSITEITGDVLLYYELSNYFQNHRHFMKS 59
V +S++E + YTD + DV L +G + L+ E+ ++Q + + KS
Sbjct 52 VYSSLMETTIPYTDDLSTDVYLP--------------SGKIYLFLEMREFYQTNLRYSKS 97
Query 60 RSDKQLEGKVYTAASEVRTACDPRVESSDGRVLHPCGLSAGSVFTDVFSAVEEDEITEIE 119
S QL G+ + A P + DG+ ++P GL S D E EI +E
Sbjct 98 ISYDQLRGE----RPKSLNAASP-LSYEDGKAIYPAGLLPNSFPHD------EYEIDGVE 146
Query 120 LDESREAICWDWDLSTFKNPSSEEINAAAPQVDFWLFNQTYASALLMDKPGVGWGVENSH 179
++ + I W+ + + + PS AP + W N T L + N
Sbjct 147 IET--DGISWESERNIVRPPSYTRDEVVAPPL--WP-NYTEVPDLSL----------NER 191
Query 180 FIVWVRESPLPVFRKVYG 197
F W+ +P P FRK++G
Sbjct 192 FTNWIYIAPFPSFRKLWG 209
> Hs22042653
Length=569
Score = 31.2 bits (69), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 17/55 (30%), Positives = 26/55 (47%), Gaps = 2/55 (3%)
Query 55 HFMKSRSDKQLEGKVYTAASEVRTACDPRVESSDG--RVLHPCGLSAGSVFTDVF 107
HF+K+ ++ EGKVY + VRT + S ++ P L+ G V F
Sbjct 102 HFLKASRSQRGEGKVYLGQASVRTLLISSINSKQFVLKLRPPKALTPGEVLVQTF 156
> At2g24950
Length=411
Score = 30.8 bits (68), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 15/42 (35%), Positives = 24/42 (57%), Gaps = 1/42 (2%)
Query 44 YELSNYFQNHRHFMKSRSDKQLEGKVYTAASEVRTACDPRVE 85
Y N N HF SR+ K++ GK+Y A+ + + DP+V+
Sbjct 165 YNQYNVDNNQHHFAVSRT-KKIIGKIYNGATMILSINDPKVK 205
> 7296376
Length=730
Score = 30.4 bits (67), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 17/51 (33%), Positives = 25/51 (49%), Gaps = 0/51 (0%)
Query 30 DTSITEITGDVLLYYELSNYFQNHRHFMKSRSDKQLEGKVYTAASEVRTAC 80
D+SI I V + YELS +KSR D +L + + ++E R A
Sbjct 427 DSSIMSIARPVDIEYELSETSDTGEQMIKSRKDSRLPSTILSQSAEPRNAA 477
Lambda K H
0.317 0.132 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3658712052
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40