bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0700_orf1
Length=143
Score E
Sequences producing significant alignments: (Bits) Value
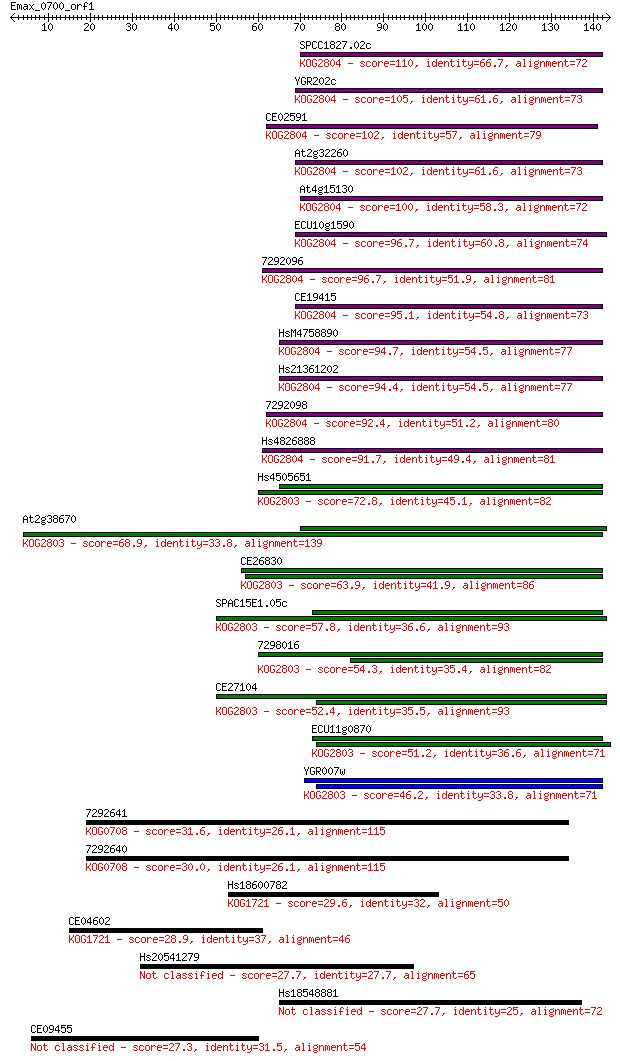
SPCC1827.02c 110 8e-25
YGR202c 105 4e-23
CE02591 102 2e-22
At2g32260 102 3e-22
At4g15130 100 9e-22
ECU10g1590 96.7 1e-20
7292096 96.7 1e-20
CE19415 95.1 4e-20
HsM4758890 94.7 5e-20
Hs21361202 94.4 6e-20
7292098 92.4 3e-19
Hs4826888 91.7 4e-19
Hs4505651 72.8 2e-13
At2g38670 68.9 3e-12
CE26830 63.9 1e-10
SPAC15E1.05c 57.8 7e-09
7298016 54.3 8e-08
CE27104 52.4 3e-07
ECU11g0870 51.2 6e-07
YGR007w 46.2 2e-05
7292641 31.6 0.49
7292640 30.0 1.6
Hs18600782 29.6 2.1
CE04602 28.9 3.8
Hs20541279 27.7 6.8
Hs18548881 27.7 7.2
CE09455 27.3 8.8
> SPCC1827.02c
Length=354
Score = 110 bits (276), Expect = 8e-25, Method: Compositional matrix adjust.
Identities = 48/72 (66%), Positives = 61/72 (84%), Gaps = 0/72 (0%)
Query 70 RPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQTVQSLEERAE 129
RPVR+Y DGV+DL H+GHMRQLEQAKK+F +V+L+ G+ +D+ THRLKG TV + +ERAE
Sbjct 92 RPVRVYADGVFDLFHIGHMRQLEQAKKVFPNVHLIVGLPNDQLTHRLKGLTVMNDKERAE 151
Query 130 TLRHIKWVDEVI 141
LRH KWVDEV+
Sbjct 152 ALRHCKWVDEVL 163
> YGR202c
Length=424
Score = 105 bits (261), Expect = 4e-23, Method: Compositional matrix adjust.
Identities = 45/73 (61%), Positives = 58/73 (79%), Gaps = 0/73 (0%)
Query 69 NRPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQTVQSLEERA 128
+RP+RIY DGV+DL HLGHM+QLEQ KK F +V L+ GV SD+ TH+LKG TV + ++R
Sbjct 101 DRPIRIYADGVFDLFHLGHMKQLEQCKKAFPNVTLIVGVPSDKITHKLKGLTVLTDKQRC 160
Query 129 ETLRHIKWVDEVI 141
ETL H +WVDEV+
Sbjct 161 ETLTHCRWVDEVV 173
> CE02591
Length=347
Score = 102 bits (254), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 45/79 (56%), Positives = 57/79 (72%), Gaps = 0/79 (0%)
Query 62 LAEGEDPNRPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQTV 121
+AE + RPVRIY DG+YDL H GH QL Q KKMF +VYL+ GV D +TH+ KG+TV
Sbjct 56 MAEANEAGRPVRIYADGIYDLFHHGHANQLRQVKKMFPNVYLIVGVCGDRDTHKYKGRTV 115
Query 122 QSLEERAETLRHIKWVDEV 140
S EER + +RH ++VDEV
Sbjct 116 TSEEERYDGVRHCRYVDEV 134
> At2g32260
Length=332
Score = 102 bits (253), Expect = 3e-22, Method: Compositional matrix adjust.
Identities = 45/74 (60%), Positives = 55/74 (74%), Gaps = 1/74 (1%)
Query 69 NRPVRIYTDGVYDLLHLGHMRQLEQAKKMF-KHVYLMAGVASDEETHRLKGQTVQSLEER 127
+RPVR+Y DG+YDL H GH R LEQAK F + YL+ G +DE TH+ KG+TV + EER
Sbjct 32 DRPVRVYADGIYDLFHFGHARSLEQAKLAFPNNTYLLVGCCNDETTHKYKGRTVMTAEER 91
Query 128 AETLRHIKWVDEVI 141
E+LRH KWVDEVI
Sbjct 92 YESLRHCKWVDEVI 105
> At4g15130
Length=298
Score = 100 bits (249), Expect = 9e-22, Method: Compositional matrix adjust.
Identities = 42/72 (58%), Positives = 54/72 (75%), Gaps = 0/72 (0%)
Query 70 RPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQTVQSLEERAE 129
RPVR+Y DG++DL H GH R +EQAKK F + YL+ G +DE T++ KG+TV + ER E
Sbjct 19 RPVRVYADGIFDLFHFGHARAIEQAKKSFPNTYLLVGCCNDEITNKFKGKTVMTESERYE 78
Query 130 TLRHIKWVDEVI 141
+LRH KWVDEVI
Sbjct 79 SLRHCKWVDEVI 90
> ECU10g1590
Length=278
Score = 96.7 bits (239), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 45/74 (60%), Positives = 52/74 (70%), Gaps = 0/74 (0%)
Query 69 NRPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQTVQSLEERA 128
NRPVRIY DGVYDL H GH R L+QAK +F +VYL+ GV D+ T RLKG V ERA
Sbjct 40 NRPVRIYCDGVYDLFHYGHARSLKQAKNLFPNVYLLVGVTDDDITVRLKGNLVMDENERA 99
Query 129 ETLRHIKWVDEVIA 142
E L H ++VDEVI
Sbjct 100 EGLIHCRYVDEVIT 113
> 7292096
Length=347
Score = 96.7 bits (239), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 42/81 (51%), Positives = 58/81 (71%), Gaps = 0/81 (0%)
Query 61 ELAEGEDPNRPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQT 120
++A R VR+Y DG+YDL H GH RQL QAK +F +VYL+ GV +DE THR+KG+T
Sbjct 16 QMARSGQTTRRVRVYADGIYDLFHQGHARQLMQAKNIFPNVYLIVGVCNDELTHRMKGRT 75
Query 121 VQSLEERAETLRHIKWVDEVI 141
V + ER E +RH ++VDE++
Sbjct 76 VMNGFERYEGVRHCRYVDEIV 96
> CE19415
Length=183
Score = 95.1 bits (235), Expect = 4e-20, Method: Compositional matrix adjust.
Identities = 40/73 (54%), Positives = 54/73 (73%), Gaps = 0/73 (0%)
Query 69 NRPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQTVQSLEERA 128
+RP+R+Y DG+YD+ H GH +QL Q K+MF VYL+ GV SDE T + KG TVQS ER
Sbjct 39 SRPIRVYADGIYDMFHYGHAKQLLQIKQMFPMVYLIVGVCSDENTLKFKGPTVQSENERY 98
Query 129 ETLRHIKWVDEVI 141
E++R ++VDEV+
Sbjct 99 ESVRQCRYVDEVL 111
> HsM4758890
Length=330
Score = 94.7 bits (234), Expect = 5e-20, Method: Compositional matrix adjust.
Identities = 42/77 (54%), Positives = 54/77 (70%), Gaps = 0/77 (0%)
Query 65 GEDPNRPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQTVQSL 124
G +RPVR+Y DG++DL H GH R L QAK +F + YL+ GV SD+ TH+ KG TV +
Sbjct 70 GTPADRPVRVYADGIFDLFHSGHARALMQAKTLFPNSYLLVGVCSDDLTHKFKGFTVMNE 129
Query 125 EERAETLRHIKWVDEVI 141
ER E LRH ++VDEVI
Sbjct 130 AERYEALRHCRYVDEVI 146
> Hs21361202
Length=369
Score = 94.4 bits (233), Expect = 6e-20, Method: Compositional matrix adjust.
Identities = 42/77 (54%), Positives = 54/77 (70%), Gaps = 0/77 (0%)
Query 65 GEDPNRPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQTVQSL 124
G +RPVR+Y DG++DL H GH R L QAK +F + YL+ GV SD+ TH+ KG TV +
Sbjct 70 GTPADRPVRVYADGIFDLFHSGHARALMQAKTLFPNSYLLVGVCSDDLTHKFKGFTVMNE 129
Query 125 EERAETLRHIKWVDEVI 141
ER E LRH ++VDEVI
Sbjct 130 AERYEALRHCRYVDEVI 146
> 7292098
Length=381
Score = 92.4 bits (228), Expect = 3e-19, Method: Compositional matrix adjust.
Identities = 41/80 (51%), Positives = 56/80 (70%), Gaps = 0/80 (0%)
Query 62 LAEGEDPNRPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQTV 121
+A R VR+Y DG+YDL H GH RQL QAK +F +VYL+ GV +DE T R+KG+TV
Sbjct 69 MARAGKTTRRVRVYADGIYDLFHQGHARQLMQAKNVFPNVYLIVGVCNDELTLRMKGRTV 128
Query 122 QSLEERAETLRHIKWVDEVI 141
+ ER E +RH ++VDE++
Sbjct 129 MNGFERYEAVRHCRYVDEIV 148
> Hs4826888
Length=367
Score = 91.7 bits (226), Expect = 4e-19, Method: Compositional matrix adjust.
Identities = 40/81 (49%), Positives = 54/81 (66%), Gaps = 0/81 (0%)
Query 61 ELAEGEDPNRPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQT 120
E + G RPVR+Y DG++DL H GH R L QAK +F + YL+ GV SDE TH KG T
Sbjct 66 EASRGTPCERPVRVYADGIFDLFHSGHARALMQAKNLFPNTYLIVGVCSDELTHNFKGFT 125
Query 121 VQSLEERAETLRHIKWVDEVI 141
V + ER + ++H ++VDEV+
Sbjct 126 VMNENERYDAVQHCRYVDEVV 146
> Hs4505651
Length=389
Score = 72.8 bits (177), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 36/77 (46%), Positives = 49/77 (63%), Gaps = 2/77 (2%)
Query 65 GEDPNRPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQTVQSL 124
G R VR++ DG YD++H GH QL QA+ M YL+ GV +DEE + KG V +
Sbjct 16 GPGGRRAVRVWCDGCYDMVHYGHSNQLRQARAMGD--YLIVGVHTDEEIAKHKGPPVFTQ 73
Query 125 EERAETLRHIKWVDEVI 141
EER + ++ IKWVDEV+
Sbjct 74 EERYKMVQAIKWVDEVV 90
Score = 53.9 bits (128), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 26/85 (30%), Positives = 50/85 (58%), Gaps = 3/85 (3%)
Query 60 VELAEGEDPN-RPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKG 118
++ A G++P IY G +DL H+GH+ LE+ ++ + Y++AG+ D+E + KG
Sbjct 201 IQFASGKEPQPGETVIYVAGAFDLFHIGHVDFLEKVHRLAERPYIIAGLHFDQEVNHYKG 260
Query 119 QT--VQSLEERAETLRHIKWVDEVI 141
+ + +L ER ++ ++V EV+
Sbjct 261 KNYPIMNLHERTLSVLACRYVSEVV 285
> At2g38670
Length=421
Score = 68.9 bits (167), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 33/73 (45%), Positives = 45/73 (61%), Gaps = 2/73 (2%)
Query 70 RPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQTVQSLEERAE 129
+PVR+Y DG +D++H GH L QA+ + L+ GV SDEE KG V L ER
Sbjct 53 KPVRVYMDGCFDMMHYGHCNALRQARALGDQ--LVVGVVSDEEIIANKGPPVTPLHERMT 110
Query 130 TLRHIKWVDEVIA 142
++ +KWVDEVI+
Sbjct 111 MVKAVKWVDEVIS 123
Score = 52.8 bits (125), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 37/140 (26%), Positives = 67/140 (47%), Gaps = 22/140 (15%)
Query 4 SELRRAQTHCAAPPYFELTSRATGNSARPALLKTSMKKRKATEDPSPQPDSQGTAPVELA 63
S L+R +H + P FE + + G L TS + + + P PD++
Sbjct 205 SSLQRQFSHGHSSPKFEDGASSAGTRV-SHFLPTSRRIVQFSNGKGPGPDAR-------- 255
Query 64 EGEDPNRPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKG--QTV 121
IY DG +DL H GH+ L +A+++ +L+ G+ +D+ +G + +
Sbjct 256 ---------IIYIDGAFDLFHAGHVEILRRARELGD--FLLVGIHNDQTVSAKRGAHRPI 304
Query 122 QSLEERAETLRHIKWVDEVI 141
+L ER+ ++ ++VDEVI
Sbjct 305 MNLHERSLSVLACRYVDEVI 324
> CE26830
Length=370
Score = 63.9 bits (154), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 36/86 (41%), Positives = 49/86 (56%), Gaps = 8/86 (9%)
Query 56 GTAPVELAEGEDPNRPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHR 115
G AP L +G R++ DG YD++H GH QL QAK+ + L+ GV +DEE
Sbjct 3 GLAPDGLPKGN------RVWADGCYDMVHFGHANQLRQAKQFGQK--LIVGVHNDEEIRL 54
Query 116 LKGQTVQSLEERAETLRHIKWVDEVI 141
KG V + +ER + IKWVDEV+
Sbjct 55 HKGPPVFNEQERYRMVAGIKWVDEVV 80
Score = 53.1 bits (126), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 32/89 (35%), Positives = 51/89 (57%), Gaps = 7/89 (7%)
Query 57 TAPVELAEGEDPNRPVR--IYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETH 114
T +E AEG P +P +Y G +DL H+GH+ LE+AK+ YL+ G+ SD+ +
Sbjct 188 TTILEFAEGRPP-KPTDKVVYVTGSFDLFHIGHLAFLEKAKEFGD--YLIVGILSDQTVN 244
Query 115 RLKGQT--VQSLEERAETLRHIKWVDEVI 141
+ KG + S+ ER ++ K V+EV+
Sbjct 245 QYKGSNHPIMSIHERVLSVLAYKPVNEVV 273
> SPAC15E1.05c
Length=365
Score = 57.8 bits (138), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 29/69 (42%), Positives = 40/69 (57%), Gaps = 2/69 (2%)
Query 73 RIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQTVQSLEERAETLR 132
R++ DG D H GH + QAK++ + L+ G+ SDEE KG V +LEER +
Sbjct 10 RLWLDGCMDFFHYGHSNAILQAKQLGET--LVIGIHSDEEITLNKGPPVMTLEERCLSAN 67
Query 133 HIKWVDEVI 141
KWVDEV+
Sbjct 68 TCKWVDEVV 76
Score = 42.4 bits (98), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 26/93 (27%), Positives = 46/93 (49%), Gaps = 8/93 (8%)
Query 50 PQPDSQGTAPVELAEGEDPNRPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVAS 109
P+ GT + L +P + + IY DG +DL H+ LE +MF + +MAG+ +
Sbjct 186 PETLISGTTLLRL----NPEKNI-IYIDGDWDLFTEKHISALELCTRMFPGIPIMAGIFA 240
Query 110 DEETHRLKGQTVQSLEERAETLRHIKWVDEVIA 142
DE+ + + +L ER L K++ ++
Sbjct 241 DEKCFE---KPMLNLLERILNLLQCKYISSILV 270
> 7298016
Length=363
Score = 54.3 bits (129), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 29/85 (34%), Positives = 51/85 (60%), Gaps = 5/85 (5%)
Query 60 VELAEGEDPNRPVRI-YTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKG 118
++ ++G+ PN +I Y G +DL H+GH+ LE+AKK+ YL+ G+ +D + KG
Sbjct 182 IQFSDGKSPNPGDKIVYVAGAFDLFHVGHLDFLEKAKKLGD--YLIVGLHTDPVVNSYKG 239
Query 119 QT--VQSLEERAETLRHIKWVDEVI 141
+ +L ER ++ K+V+EV+
Sbjct 240 SNYPIMNLHERVLSVLACKFVNEVV 264
Score = 50.1 bits (118), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 25/60 (41%), Positives = 37/60 (61%), Gaps = 2/60 (3%)
Query 82 LLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQTVQSLEERAETLRHIKWVDEVI 141
++H GH L QAK + V + G+ +DEE + KG V + EER + ++ IKWVDEV+
Sbjct 1 MVHFGHANSLRQAKALGDKV--IVGIHTDEEITKHKGPPVFTEEERVKMVKGIKWVDEVV 58
> CE27104
Length=302
Score = 52.4 bits (124), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 33/93 (35%), Positives = 48/93 (51%), Gaps = 8/93 (8%)
Query 50 PQPDSQGTAPVELAEGEDPNRPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVAS 109
P+ QG P +GE + R+YTDG +D +H + R L AK+ K L+ G+ S
Sbjct 2 PKAIVQGLTP----DGE--KKKARVYTDGCFDFVHFANARLLWPAKQYGKK--LIVGIHS 53
Query 110 DEETHRLKGQTVQSLEERAETLRHIKWVDEVIA 142
D+E + + EER + I+WVDE IA
Sbjct 54 DDELDNNGILPIFTDEERYRLISAIRWVDEEIA 86
Score = 50.8 bits (120), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 26/71 (36%), Positives = 38/71 (53%), Gaps = 4/71 (5%)
Query 74 IYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQT--VQSLEERAETL 131
+Y G +DL H GH+ LE AK + YL+ G+ D++ + KG V +L ER +
Sbjct 151 VYVSGAFDLFHAGHLSFLEAAKDLGD--YLIVGIVGDDDVNEEKGTIFPVMNLLERTLNI 208
Query 132 RHIKWVDEVIA 142
+K VDEV
Sbjct 209 SSLKIVDEVFV 219
> ECU11g0870
Length=322
Score = 51.2 bits (121), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 26/69 (37%), Positives = 40/69 (57%), Gaps = 2/69 (2%)
Query 73 RIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQTVQSLEERAETLR 132
+++ DG +D+ H GH L Q+K + YL+AGV S ++ KG V EER E +
Sbjct 8 KVWADGCFDMFHYGHANALRQSKALGD--YLIAGVHSSLSINQEKGLPVMEDEERYEVVE 65
Query 133 HIKWVDEVI 141
++VDEV+
Sbjct 66 GCRYVDEVV 74
Score = 45.1 bits (105), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 25/71 (35%), Positives = 38/71 (53%), Gaps = 3/71 (4%)
Query 74 IYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRL-KGQTVQSLEERAETLR 132
++ DG +DL H GH+ L A+ M YL+ G+ DE T + V S +ER TL
Sbjct 180 VFMDGNFDLFHAGHVASLRIARGMGD--YLIVGIHDDETTKEYTRSYPVLSTKERMLTLM 237
Query 133 HIKWVDEVIAP 143
++VDE++
Sbjct 238 ACRYVDEIVVS 248
> YGR007w
Length=323
Score = 46.2 bits (108), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 24/73 (32%), Positives = 38/73 (52%), Gaps = 2/73 (2%)
Query 71 PVRIYTDGVYDLLHLGHMRQLEQAKKMF--KHVYLMAGVASDEETHRLKGQTVQSLEERA 128
P +++ DG +D H GH + QA++ ++ L GV +DE+ KG V + ER
Sbjct 7 PDKVWIDGCFDFTHHGHAGAILQARRTVSKENGKLFCGVHTDEDIQHNKGTPVMNSSERY 66
Query 129 ETLRHIKWVDEVI 141
E R +W EV+
Sbjct 67 EHTRSNRWCSEVV 79
Score = 37.7 bits (86), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 20/69 (28%), Positives = 38/69 (55%), Gaps = 6/69 (8%)
Query 74 IYTDGVYDLLHLGHMRQLEQAK-KMFKHVYLMAGVASDEETHRLKGQTVQSLEERAETLR 132
+Y DG +DL H+G + QL + K + L+ G+ + + + T+ +++ER ++
Sbjct 200 VYVDGDFDLFHMGDIDQLRKLKMDLHPDKKLIVGITTSDYS-----STIMTMKERVLSVL 254
Query 133 HIKWVDEVI 141
K+VD VI
Sbjct 255 SCKYVDAVI 263
> 7292641
Length=158
Score = 31.6 bits (70), Expect = 0.49, Method: Compositional matrix adjust.
Identities = 30/115 (26%), Positives = 48/115 (41%), Gaps = 12/115 (10%)
Query 19 FELTSRATGNSARPALLKTSMKKRKATEDPSPQPDSQGTAPVELAEGEDPNRPVRIYTDG 78
+E R + N RP ++ +K R + S PD G+ RP R Y
Sbjct 22 YEAVQRLSINYTRPVIILGPLKDRINDDLISEYPDKFGSCVPHTT------RPKREYEVD 75
Query 79 VYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQTVQSLEERAETLRH 133
D + Q+E + + H+++ AG +D L G +V S+ E AE +H
Sbjct 76 GRDYHFVSSREQME--RDIQNHLFIEAGQYND----NLYGTSVASVREVAEKGKH 124
> 7292640
Length=960
Score = 30.0 bits (66), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 30/115 (26%), Positives = 48/115 (41%), Gaps = 12/115 (10%)
Query 19 FELTSRATGNSARPALLKTSMKKRKATEDPSPQPDSQGTAPVELAEGEDPNRPVRIYTDG 78
+E R + N RP ++ +K R + S PD G+ RP R Y
Sbjct 759 YEAVQRLSINYTRPVIILGPLKDRINDDLISEYPDKFGSCVPHTT------RPKREYEVD 812
Query 79 VYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQTVQSLEERAETLRH 133
D + Q+E + + H+++ AG +D L G +V S+ E AE +H
Sbjct 813 GRDYHFVSSREQME--RDIQNHLFIEAGQYNDN----LYGTSVASVREVAEKGKH 861
> Hs18600782
Length=453
Score = 29.6 bits (65), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 16/50 (32%), Positives = 22/50 (44%), Gaps = 5/50 (10%)
Query 53 DSQGTAPVELAEGEDPNRPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVY 102
DSQ + E+ E EDP P R+ Y H H R + +F+ Y
Sbjct 344 DSQDHSAFEMEEEEDPRAPPRL-----YGFAHPSHCRSAASKRNIFRTKY 388
> CE04602
Length=594
Score = 28.9 bits (63), Expect = 3.8, Method: Composition-based stats.
Identities = 17/46 (36%), Positives = 25/46 (54%), Gaps = 1/46 (2%)
Query 15 APPYFELTSRATGNSARPALLKTSMKKRKATEDPSPQPDSQGTAPV 60
+PP F +TS AT N++ KTS A +P P S+ T+P+
Sbjct 90 SPPPF-MTSPATSNNSVSVARKTSAPPGFAQYEPDTAPPSRSTSPL 134
> Hs20541279
Length=773
Score = 27.7 bits (60), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 18/65 (27%), Positives = 27/65 (41%), Gaps = 1/65 (1%)
Query 32 PALLKTSMKKRKATEDPSPQPDSQGTAPVELAEGEDPNRPVRIYTDGVYDLLHLGHMRQL 91
P LL ++ R + P P P +Q P L +P DG+ +L +G +
Sbjct 561 PGLLLQALGNRFCSSGPEPSPPNQ-DKPGSLGSATEPGASESSSADGMLKVLRVGMKSKW 619
Query 92 EQAKK 96
QA K
Sbjct 620 MQAWK 624
> Hs18548881
Length=549
Score = 27.7 bits (60), Expect = 7.2, Method: Composition-based stats.
Identities = 18/72 (25%), Positives = 30/72 (41%), Gaps = 2/72 (2%)
Query 65 GEDPNRPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQTVQSL 124
G+ P+ +Y + L +L H +L + ++ + S+ T R K +Q L
Sbjct 392 GQSAEPPISVYF--LPALSYLPHQLRLTCMNSSLNSMSCLSQLMSESATCRAKSSLIQGL 449
Query 125 EERAETLRHIKW 136
EE E R W
Sbjct 450 EENEERRRLFAW 461
> CE09455
Length=382
Score = 27.3 bits (59), Expect = 8.8, Method: Compositional matrix adjust.
Identities = 17/60 (28%), Positives = 24/60 (40%), Gaps = 6/60 (10%)
Query 6 LRRAQTHCAAP------PYFELTSRATGNSARPALLKTSMKKRKATEDPSPQPDSQGTAP 59
LR +T P P F + + + +P L+ MKK + T PS P AP
Sbjct 295 LRSEETEIKVPSNLSVNPIFSTNATTSSQTTKPKLIYNEMKKPRTTLKPSTAPPESSKAP 354
Lambda K H
0.314 0.129 0.372
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1639544670
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40