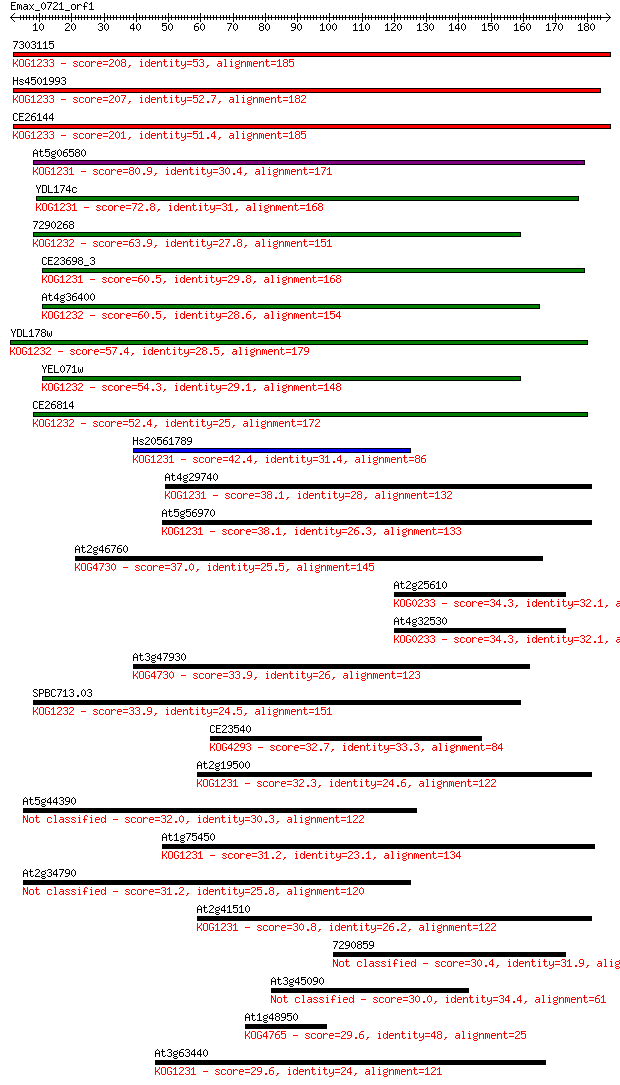
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0721_orf1
Length=186
Score E
Sequences producing significant alignments: (Bits) Value
7303115 208 4e-54
Hs4501993 207 1e-53
CE26144 201 7e-52
At5g06580 80.9 2e-15
YDL174c 72.8 4e-13
7290268 63.9 2e-10
CE23698_3 60.5 2e-09
At4g36400 60.5 2e-09
YDL178w 57.4 2e-08
YEL071w 54.3 2e-07
CE26814 52.4 6e-07
Hs20561789 42.4 6e-04
At4g29740 38.1 0.010
At5g56970 38.1 0.011
At2g46760 37.0 0.021
At2g25610 34.3 0.17
At4g32530 34.3 0.17
At3g47930 33.9 0.20
SPBC713.03 33.9 0.21
CE23540 32.7 0.40
At2g19500 32.3 0.58
At5g44390 32.0 0.74
At1g75450 31.2 1.2
At2g34790 31.2 1.4
At2g41510 30.8 1.7
7290859 30.4 1.9
At3g45090 30.0 3.1
At1g48950 29.6 3.3
At3g63440 29.6 4.1
> 7303115
Length=631
Score = 208 bits (530), Expect = 4e-54, Method: Compositional matrix adjust.
Identities = 98/185 (52%), Positives = 133/185 (71%), Gaps = 0/185 (0%)
Query 2 IVDCAIKHNICLIPFGGGTSVTLGVCTPEEEERMVATVSLSFMQKILYLDRDALLMWVEA 61
+V A KHN+ L+PFGGGTSV+ + P+ E RM+ + S M ++L+L+R+ L + E+
Sbjct 174 LVRLANKHNVMLVPFGGGTSVSGAITCPQNESRMICALDTSQMNRLLWLNRENLTVCFES 233
Query 62 GAVGASIEERLRPYGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYGNIEDLVVDIKVVT 121
G VG +E LR G+T+GHEPDS EFST+GGWVATRASGMKKN YGNIEDLVV +++VT
Sbjct 234 GIVGQDLERVLRSEGLTVGHEPDSYEFSTLGGWVATRASGMKKNVYGNIEDLVVRVRMVT 293
Query 122 TRGCMHACNSSPRISSGPSLQQLFLGSEGIYGIITQVLLKIKPIPEYKVYDCLAFPSFSL 181
G + S+PR+S GP + LGSEG G+IT+V+LK++P+P + Y LAFP+F
Sbjct 294 PSGTLERECSAPRVSCGPDFNHVILGSEGTLGVITEVVLKVRPLPSLRRYGSLAFPNFEQ 353
Query 182 GVLFL 186
GVLF+
Sbjct 354 GVLFM 358
> Hs4501993
Length=658
Score = 207 bits (526), Expect = 1e-53, Method: Compositional matrix adjust.
Identities = 96/182 (52%), Positives = 129/182 (70%), Gaps = 0/182 (0%)
Query 2 IVDCAIKHNICLIPFGGGTSVTLGVCTPEEEERMVATVSLSFMQKILYLDRDALLMWVEA 61
IV+ A K+N+C+IP GGGTSV+ G+ P +E R + ++ S M +IL++D + L VEA
Sbjct 221 IVNLACKYNLCIIPIGGGTSVSYGLMCPADETRTIISLDTSQMNRILWVDENNLTAHVEA 280
Query 62 GAVGASIEERLRPYGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYGNIEDLVVDIKVVT 121
G G +E +L+ G GHEPDS+EFSTVGGWV+TRASGMKKN YGNIEDLVV IK+VT
Sbjct 281 GITGQELERQLKESGYCTGHEPDSLEFSTVGGWVSTRASGMKKNIYGNIEDLVVHIKMVT 340
Query 122 TRGCMHACNSSPRISSGPSLQQLFLGSEGIYGIITQVLLKIKPIPEYKVYDCLAFPSFSL 181
RG + PR+S+GP + +GSEG G+IT+ +KI+P+PEY+ Y +AFP+F
Sbjct 341 PRGIIEKSCQGPRMSTGPDIHHFIMGSEGTLGVITEATIKIRPVPEYQKYGSVAFPNFEQ 400
Query 182 GV 183
GV
Sbjct 401 GV 402
> CE26144
Length=597
Score = 201 bits (511), Expect = 7e-52, Method: Compositional matrix adjust.
Identities = 95/185 (51%), Positives = 129/185 (69%), Gaps = 0/185 (0%)
Query 2 IVDCAIKHNICLIPFGGGTSVTLGVCTPEEEERMVATVSLSFMQKILYLDRDALLMWVEA 61
I++ A+ HN +IP GGGTSVT + TPE E+R V ++ ++ + KIL++DR+ L +A
Sbjct 150 IIEGAMSHNCAIIPIGGGTSVTNALDTPETEKRAVISMDMALLDKILWIDRENLTCRAQA 209
Query 62 GAVGASIEERLRPYGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYGNIEDLVVDIKVVT 121
G VG S+E +L G T GHEPDS+EFST+GGWV+TRASGMKKN+YGNIEDLVV + V
Sbjct 210 GIVGQSLERQLNKKGFTCGHEPDSIEFSTLGGWVSTRASGMKKNKYGNIEDLVVHLNFVC 269
Query 122 TRGCMHACNSSPRISSGPSLQQLFLGSEGIYGIITQVLLKIKPIPEYKVYDCLAFPSFSL 181
+G + PR+SSGP + Q+ LGSEG G++++V +KI PIPE K + FP+F
Sbjct 270 PKGIIQKQCQVPRMSSGPDIHQIILGSEGTLGVVSEVTIKIFPIPEVKRFGSFVFPNFES 329
Query 182 GVLFL 186
GV F
Sbjct 330 GVNFF 334
> At5g06580
Length=417
Score = 80.9 bits (198), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 52/172 (30%), Positives = 90/172 (52%), Gaps = 7/172 (4%)
Query 8 KHNICLIPFGGGTSVTLGVCTPEEEERMVATVSLSFMQKILYLDRDALLMWVEAGAVGAS 67
++ + ++P+GG TS+ P + + +S M+++ L + + + VE G
Sbjct 17 EYKVPIVPYGGATSIEGHTLAP----KGGVCIDMSLMKRVKALHVEDMDVIVEPGIGWLE 72
Query 68 IEERLRPYGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYGNIEDLVVDIKVVTTRGCMH 127
+ E L YG+ +P +++GG ATR SG RYG + D V+ +KVV G +
Sbjct 73 LNEYLEEYGLFFPLDPGP--GASIGGMCATRCSGSLAVRYGTMRDNVISLKVVLPNGDVV 130
Query 128 ACNSSPRISS-GPSLQQLFLGSEGIYGIITQVLLKIKPIPEYKVYDCLAFPS 178
S R S+ G L +L +GSEG G+IT++ L+++ IP++ V FP+
Sbjct 131 KTASRARKSAAGYDLTRLIIGSEGTLGVITEITLRLQKIPQHSVVAVCNFPT 182
> YDL174c
Length=587
Score = 72.8 bits (177), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 52/170 (30%), Positives = 89/170 (52%), Gaps = 6/170 (3%)
Query 9 HNICLIPFGGGTSVTLGVCTPEEEERMVATVSLS-FMQKILYLDRDALLMWVEAGAVGAS 67
+N+ ++PF GGTS+ G P + TV LS FM ++ D+ L + V+AG
Sbjct 173 NNMPVVPFSGGTSLE-GHFLPTRIGDTI-TVDLSKFMNNVVKFDKLDLDITVQAGLPWED 230
Query 68 IEERLRPYGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYGNIEDLVVDIKVVTTRGCMH 127
+ + L +G+ G +P + +GG +A SG RYG +++ ++++ +V G +
Sbjct 231 LNDYLSDHGLMFGCDPGPG--AQIGGCIANSCSGTNAYRYGTMKENIINMTIVLPDGTIV 288
Query 128 ACNSSPRISS-GPSLQQLFLGSEGIYGIITQVLLKIKPIPEYKVYDCLAF 176
PR SS G +L LF+GSEG GI+T+ +K P+ + ++F
Sbjct 289 KTKKRPRKSSAGYNLNGLFVGSEGTLGIVTEATVKCHVKPKAETVAVVSF 338
> 7290268
Length=533
Score = 63.9 bits (154), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 42/152 (27%), Positives = 84/152 (55%), Gaps = 5/152 (3%)
Query 8 KHNICLIPFGGGTSVTLGVCTPEEEERMVATVSLSFMQKILYLDRDALLMWVEAGAVGAS 67
+ + ++P GG T + +G P +E +SL+ + K+L +D + VEAG + +
Sbjct 135 ERRLAVVPQGGNTGL-VGGSVPICDE---IVLSLARLNKVLSVDEVTGIAVVEAGCILEN 190
Query 68 IEERLRPYGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYGNIEDLVVDIKVVTTRG-CM 126
++R R G+T+ + + +GG V+T A G++ RYGN+ V+ ++ V G +
Sbjct 191 FDQRAREVGLTVPLDLGAKASCHIGGNVSTNAGGVRVVRYGNLHGSVLGVEAVLATGQVL 250
Query 127 HACNSSPRISSGPSLQQLFLGSEGIYGIITQV 158
++ + ++G ++ LF+GSEG G++T++
Sbjct 251 DLMSNFKKDNTGYHMKHLFIGSEGTLGVVTKL 282
> CE23698_3
Length=454
Score = 60.5 bits (145), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 50/174 (28%), Positives = 86/174 (49%), Gaps = 14/174 (8%)
Query 11 ICLIPFGGGTSVTLGVCTPEEEERMVATVSLSFMQKI--LYLDRDALLMWVEAGAVGASI 68
I ++PFG GT + G + + + +S Q I L + V+ ++
Sbjct 56 IPVVPFGTGTGLEGGSMS------TLGGICISTQQIIGDPVLREQDFVCSVKPSTTRIAL 109
Query 69 EERLRPYGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYGNIEDLVVDIKVVTTRGCM-- 126
+ ++ G+ +P + ++V G VAT ASG RYG +++ VV+++VV G +
Sbjct 110 NDAIKNSGLFFPVDPGAD--ASVCGMVATSASGTNAIRYGTMKENVVNLEVVLADGTIID 167
Query 127 -HACNSSPRISS-GPSLQQLFLGSEGIYGIITQVLLKIKPIPEYKVYDCLAFPS 178
PR SS G + +LF+GSEG GIIT+ +K+ P P++ +FP+
Sbjct 168 TKGKGRCPRKSSAGFNFTELFVGSEGTLGIITEATVKVHPRPQFLSAAVCSFPT 221
> At4g36400
Length=524
Score = 60.5 bits (145), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 44/155 (28%), Positives = 81/155 (52%), Gaps = 5/155 (3%)
Query 11 ICLIPFGGGTSVTLGVCTPEEEERMVATVSLSFMQKILYLDRDALLMWVEAGAVGASIEE 70
+ ++P GG T + +G P +E V++ M KIL D + ++ EAG + ++
Sbjct 126 LAVVPQGGNTGL-VGGSVPVFDE---VIVNVGLMNKILSFDEVSGVLVCEAGCILENLAT 181
Query 71 RLRPYGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYGNIEDLVVDIKVVTTRG-CMHAC 129
L G + + + +GG V+T A G++ RYG++ V+ ++ VT G +
Sbjct 182 FLDTKGFIMPLDLGAKGSCHIGGNVSTNAGGLRLIRYGSLHGTVLGLEAVTANGNVLDML 241
Query 130 NSSPRISSGPSLQQLFLGSEGIYGIITQVLLKIKP 164
+ + ++G L+ LF+GSEG GI+T+V + +P
Sbjct 242 GTLRKDNTGYDLKHLFIGSEGSLGIVTKVSILTQP 276
> YDL178w
Length=530
Score = 57.4 bits (137), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 51/180 (28%), Positives = 91/180 (50%), Gaps = 5/180 (2%)
Query 1 VIVDCAIKHNICLIPFGGGTSVTLGVCTPEEEERMVATVSLSFMQKILYLDRDALLMWVE 60
+I++ I ++P GG T + +G P +E ++ SL+ + KI D + ++ +
Sbjct 116 LILNYCNDEKIAVVPQGGNTGL-VGGSVPIFDELIL---SLANLNKIRDFDPVSGILKCD 171
Query 61 AGAVGASIEERLRPYGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYGNIEDLVVDIKVV 120
AG + + + + + VGG VAT A G++ RYG++ V+ ++VV
Sbjct 172 AGVILENANNYVMEQNYMFPLDLGAKGSCHVGGVVATNAGGLRLLRYGSLHGSVLGLEVV 231
Query 121 TTRG-CMHACNSSPRISSGPSLQQLFLGSEGIYGIITQVLLKIKPIPEYKVYDCLAFPSF 179
G +++ +S + ++G L+QLF+GSEG GIIT V + P P+ L+ SF
Sbjct 232 MPNGQIVNSMHSMRKDNTGYDLKQLFIGSEGTIGIITGVSILTVPKPKAFNVSYLSVESF 291
> YEL071w
Length=496
Score = 54.3 bits (129), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 43/149 (28%), Positives = 73/149 (48%), Gaps = 5/149 (3%)
Query 11 ICLIPFGGGTSVTLGVCTPEEEERMVATVSLSFMQKILYLDRDALLMWVEAGAVGASIEE 70
+ ++P GG T + +G P +E +SL M K+ D + +AG V +
Sbjct 92 LAVVPQGGNTDL-VGASVPVFDE---IVLSLRNMNKVRDFDPVSGTFKCDAGVVMRDAHQ 147
Query 71 RLRPYGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYGNIEDLVVDIKVVTTRG-CMHAC 129
L + + S VGG V+T A G+ RYG++ V+ ++VV G +
Sbjct 148 FLHDHDHIFPLDLPSRNNCQVGGVVSTNAGGLNFLRYGSLHGNVLGLEVVLPNGEIISNI 207
Query 130 NSSPRISSGPSLQQLFLGSEGIYGIITQV 158
N+ + ++G L+QLF+G+EG G++T V
Sbjct 208 NALRKDNTGYDLKQLFIGAEGTIGVVTGV 236
> CE26814
Length=487
Score = 52.4 bits (124), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 43/175 (24%), Positives = 82/175 (46%), Gaps = 7/175 (4%)
Query 8 KHNICLIPFGGGTSVTLGVCTPEEEERMVATVSLSFMQKILYLDRDALLMWVEAGAVGAS 67
K+ + ++P GG T + +G P +E +S++ + K D ++ ++G +
Sbjct 88 KNKLAVVPQGGNTGL-VGGSIPVHDE---VVISMNKINKQFSFDDTMGILKCDSGFILED 143
Query 68 IEERLRPYGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYGNIEDLVVDIKVVTTR---G 124
++ +L G + + + +GG +AT A G++ RYG++ ++ + VV
Sbjct 144 LDNKLAKLGYMMPFDLGAKGSCQIGGNIATCAGGIRLIRYGSLHAHLLGLTVVLPDEHGT 203
Query 125 CMHACNSSPRISSGPSLQQLFLGSEGIYGIITQVLLKIKPIPEYKVYDCLAFPSF 179
+H +S + ++ LFLGSEG G+IT V + P P+ L SF
Sbjct 204 VLHLGSSIRKDNTTLHTPHLFLGSEGQLGVITSVTMTAVPKPKSVQSAMLGIESF 258
> Hs20561789
Length=230
Score = 42.4 bits (98), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 27/86 (31%), Positives = 47/86 (54%), Gaps = 2/86 (2%)
Query 39 VSLSFMQKILYLDRDALLMWVEAGAVGASIEERLRPYGVTLGHEPDSMEFSTVGGWVATR 98
V+L+ M +IL L+++ + VE G ++ LR G+ +P + +++ G AT
Sbjct 134 VNLTHMDRILELNQEDFSVVVEPGVTRKALNAHLRDSGLWFPVDPGA--DASLCGMAATG 191
Query 99 ASGMKKNRYGNIEDLVVDIKVVTTRG 124
ASG RYG + D V++++VV G
Sbjct 192 ASGTNAVRYGTMRDNVLNLEVVLPDG 217
> At4g29740
Length=524
Score = 38.1 bits (87), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 37/132 (28%), Positives = 62/132 (46%), Gaps = 12/132 (9%)
Query 49 YLDRDALLMWVEAGAVGASIEERLRPYGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYG 108
Y D A MWV+ + A+++ + P T D + S VGG ++ G + R+G
Sbjct 145 YADVAAGTMWVDV--LKAAVDRGVSPVTWT-----DYLYLS-VGGTLSNAGIGGQTFRHG 196
Query 109 NIEDLVVDIKVVTTRGCMHACNSSPRISSGPSLQQLFLGSEGIYGIITQVLLKIKPIPEY 168
V ++ V+T +G M C SP+++ P L LG G +GIIT+ + + P
Sbjct 197 PQISNVHELDVITGKGEMMTC--SPKLN--PELFYGVLGGLGQFGIITRARIALDHAPTR 252
Query 169 KVYDCLAFPSFS 180
+ + + FS
Sbjct 253 VKWSRILYSDFS 264
> At5g56970
Length=523
Score = 38.1 bits (87), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 35/133 (26%), Positives = 60/133 (45%), Gaps = 12/133 (9%)
Query 48 LYLDRDALLMWVEAGAVGASIEERLRPYGVTLGHEPDSMEFSTVGGWVATRASGMKKNRY 107
LY+D DA +W+E + ++E L P T + TVGG ++ + RY
Sbjct 138 LYVDVDAAWLWIEV--LNKTLELGLTPVSWT------DYLYLTVGGTLSNGGISGQTFRY 189
Query 108 GNIEDLVVDIKVVTTRGCMHACNSSPRISSGPSLQQLFLGSEGIYGIITQVLLKIKPIPE 167
G V+++ V+T +G + C+ L LG G +GIIT+ +K++ P+
Sbjct 190 GPQITNVLEMDVITGKGEIATCSK----DMNSDLFFAVLGGLGQFGIITRARIKLEVAPK 245
Query 168 YKVYDCLAFPSFS 180
+ + FS
Sbjct 246 RAKWLRFLYIDFS 258
> At2g46760
Length=603
Score = 37.0 bits (84), Expect = 0.021, Method: Compositional matrix adjust.
Identities = 37/149 (24%), Positives = 71/149 (47%), Gaps = 9/149 (6%)
Query 21 SVTLGVCTPEEEERMVATVSLSFMQKILYLDRDALLMWVEAGAVGASIEERLRPYGVTLG 80
S+T CT + +++T F+ + D A+ + VE+G + G+ L
Sbjct 103 SITKLACTDGTDGLLIST---KFLNHTVRTDATAMTLTVESGVTLRQLIAEAAKVGLALP 159
Query 81 HEPDSMEFSTVGGWVATRASGMKKNRYGN-IEDLVVDIKVVTTRGCMHACNSSPRISSGP 139
+ P TVGG + T A G G+ + D V +I++V+ G ++ + R+
Sbjct 160 YAPYWWGL-TVGGMMGTGAHGSSLWGKGSAVHDYVTEIRIVSP-GSVNDGFAKVRVLRET 217
Query 140 SLQQLFLGSE---GIYGIITQVLLKIKPI 165
+ + F ++ G+ G+I+QV LK++P+
Sbjct 218 TTPKEFNAAKVSLGVLGVISQVTLKLQPM 246
> At2g25610
Length=178
Score = 34.3 bits (77), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 17/55 (30%), Positives = 31/55 (56%), Gaps = 2/55 (3%)
Query 120 VTTRGCMHACNSSPRISSGPSLQQLFLGSEGIYGIITQVLL--KIKPIPEYKVYD 172
+T + A +PRI+S + +F + IYG+I ++L K++ +P K+YD
Sbjct 45 ITGSSLIGAAIEAPRITSKNLISVIFCEAVAIYGVIVAIILQTKLESVPSSKMYD 99
> At4g32530
Length=180
Score = 34.3 bits (77), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 17/55 (30%), Positives = 31/55 (56%), Gaps = 2/55 (3%)
Query 120 VTTRGCMHACNSSPRISSGPSLQQLFLGSEGIYGIITQVLL--KIKPIPEYKVYD 172
+T + A +PRI+S + +F + IYG+I ++L K++ +P K+YD
Sbjct 47 ITGSSLIGAAIEAPRITSKNLISVIFCEAVAIYGVIVAIILQTKLESVPSSKMYD 101
> At3g47930
Length=610
Score = 33.9 bits (76), Expect = 0.20, Method: Compositional matrix adjust.
Identities = 32/124 (25%), Positives = 60/124 (48%), Gaps = 8/124 (6%)
Query 39 VSLSFMQKILYLDRDALLMWVEAGAVGASIEERLRPYGVTLGHEPDSMEFSTVGGWVATR 98
V+L+ M K+L +D++ + V+AG + + ++ YG+TL + S+ +GG +
Sbjct 169 VNLALMDKVLEVDKEKKRVTVQAGIRVQQLVDAIKDYGLTLQNFA-SIREQQIGGIIQVG 227
Query 99 ASGMKKNRYGNIEDLVVDIKVVT-TRGCMHACNSSPRISSGPSLQQLFLGSEGIYGIITQ 157
A G R I++ V+ +K+VT +G + P L L G G++ +
Sbjct 228 AHGTGA-RLPPIDEQVISMKLVTPAKGTIELSR-----EKDPELFHLARCGLGGLGVVAE 281
Query 158 VLLK 161
V L+
Sbjct 282 VTLQ 285
> SPBC713.03
Length=526
Score = 33.9 bits (76), Expect = 0.21, Method: Compositional matrix adjust.
Identities = 37/152 (24%), Positives = 77/152 (50%), Gaps = 5/152 (3%)
Query 8 KHNICLIPFGGGTSVTLGVCTPEEEERMVATVSLSFMQKILYLDRDALLMWVEAGAVGAS 67
+ + ++P GG T + +G P +E ++L M +I D + ++ +++G + +
Sbjct 118 QKKLAVVPQGGNTGL-VGGSVPVFDE---IVLNLGLMNQIHTFDEISGVITLDSGVILEN 173
Query 68 IEERLRPYGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYGNIEDLVVDIKVVTTRGC-M 126
+ L G + + VGG AT A G++ RYG++ ++ ++ V G +
Sbjct 174 ADNFLAEKGYMFPLDLGAKGSCQVGGCAATAAGGLRLLRYGSLHGSILGMEAVLPDGTIL 233
Query 127 HACNSSPRISSGPSLQQLFLGSEGIYGIITQV 158
+ + ++G ++QLF+GSEG G+IT++
Sbjct 234 DNLVTLRKDNTGLDIKQLFIGSEGYLGVITKL 265
> CE23540
Length=1002
Score = 32.7 bits (73), Expect = 0.40, Method: Compositional matrix adjust.
Identities = 28/91 (30%), Positives = 40/91 (43%), Gaps = 7/91 (7%)
Query 63 AVGASIEERLRPYGV----TLGHEPDSMEFSTVGGWVATRASG---MKKNRYGNIEDLVV 115
A+G S + ++ P V +LG +P SM+FST G+ R SG ++ N E V
Sbjct 618 ALGFSNDGKMNPANVIECSSLGSQPLSMKFSTNSGYSNDRISGEEAIRSQYITNTETSYV 677
Query 116 DIKVVTTRGCMHACNSSPRISSGPSLQQLFL 146
D K+ NS+ I QQ L
Sbjct 678 DGKIYCKGTVRSDGNSNAAIFKYTPKQQYHL 708
> At2g19500
Length=515
Score = 32.3 bits (72), Expect = 0.58, Method: Compositional matrix adjust.
Identities = 30/122 (24%), Positives = 55/122 (45%), Gaps = 5/122 (4%)
Query 59 VEAGAVGASIEERLRPYGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYGNIEDLVVDIK 118
V AG + + ++ GV+ D + TVGG ++ G + R G + V+++
Sbjct 125 VAAGTLWVDVLKKTAEKGVSPVSWTDYLHI-TVGGTLSNGGIGGQVFRNGPLVSNVLELD 183
Query 119 VVTTRGCMHACNSSPRISSGPSLQQLFLGSEGIYGIITQVLLKIKPIPEYKVYDCLAFPS 178
V+T +G M C+ P L LG G +GIIT+ + + P+ + + +
Sbjct 184 VITGKGEMLTCSR----QLNPELFYGVLGGLGQFGIITRARIVLDHAPKRAKWFRMLYSD 239
Query 179 FS 180
F+
Sbjct 240 FT 241
> At5g44390
Length=542
Score = 32.0 bits (71), Expect = 0.74, Method: Compositional matrix adjust.
Identities = 37/126 (29%), Positives = 54/126 (42%), Gaps = 10/126 (7%)
Query 5 CAIKHNICLIPFGGGTSVTLGVCTPEEEERMVATVSLSFMQKILYLDRDALLMWVEAGA- 63
C+ K I GG GV + E+ + LS +++I +D WVEAGA
Sbjct 104 CSKKLEIHFRVRSGGHDYE-GVSYVSQIEKPFVLIDLSKLRQI-NVDIKDTSAWVEAGAT 161
Query 64 VGA---SIEERLRPYGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYGNIEDLVVDIKVV 120
VG I E+ + +G G P +GG + A G +YG D V+D K+V
Sbjct 162 VGELYYRIAEKSKFHGFPAGVYPSL----GIGGHITGGAYGSLMRKYGLAADNVLDAKIV 217
Query 121 TTRGCM 126
G +
Sbjct 218 DANGKL 223
> At1g75450
Length=536
Score = 31.2 bits (69), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 31/134 (23%), Positives = 59/134 (44%), Gaps = 12/134 (8%)
Query 48 LYLDRDALLMWVEAGAVGASIEERLRPYGVTLGHEPDSMEFSTVGGWVATRASGMKKNRY 107
+Y+D +WV+ + ++E L P T + TVGG ++ + +
Sbjct 132 MYVDVWGGELWVDV--LKKTLEHGLAPKSWT------DYLYLTVGGTLSNAGISGQAFHH 183
Query 108 GNIEDLVVDIKVVTTRGCMHACNSSPRISSGPSLQQLFLGSEGIYGIITQVLLKIKPIPE 167
G V+++ VVT +G + C+ L LG G +GIIT+ + ++P P+
Sbjct 184 GPQISNVLELDVVTGKGEVMRCSEE----ENTRLFHGVLGGLGQFGIITRARISLEPAPQ 239
Query 168 YKVYDCLAFPSFSL 181
+ + + SF +
Sbjct 240 RVRWIRVLYSSFKV 253
> At2g34790
Length=532
Score = 31.2 bits (69), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 31/120 (25%), Positives = 50/120 (41%), Gaps = 2/120 (1%)
Query 5 CAIKHNICLIPFGGGTSVTLGVCTPEEEERMVATVSLSFMQKILYLDRDALLMWVEAGAV 64
CA K + L GG G+ E+E V LS ++++ +D D+ W AGA
Sbjct 100 CAKKLQLHLRLRSGGHDYE-GLSFVAEDETPFVIVDLSKLRQV-DVDLDSNSAWAHAGAT 157
Query 65 GASIEERLRPYGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYGNIEDLVVDIKVVTTRG 124
+ R++ T G +GG + A G ++G D V+D ++V G
Sbjct 158 IGEVYYRIQEKSQTHGFPAGLCSSLGIGGHLVGGAYGSMMRKFGLGADNVLDARIVDANG 217
> At2g41510
Length=575
Score = 30.8 bits (68), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 32/125 (25%), Positives = 57/125 (45%), Gaps = 11/125 (8%)
Query 59 VEAGAVGASIEERLRPYGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYGNIEDLVVDIK 118
V G + +I YG++ D + TVGG ++ + ++G + V ++
Sbjct 161 VSGGEIWINILRETLKYGLSPKSWTDYLHL-TVGGTLSNAGISGQAFKHGPQINNVYQLE 219
Query 119 VVTTRGCMHACNSSPRISSGPSLQQLF---LGSEGIYGIITQVLLKIKPIPEYKVYDCLA 175
+VT +G + C S R S +LF LG G +GIIT+ + ++P P + +
Sbjct 220 IVTGKGEVVTC-SEKRNS------ELFFSVLGGLGQFGIITRARISLEPAPHMVKWIRVL 272
Query 176 FPSFS 180
+ FS
Sbjct 273 YSDFS 277
> 7290859
Length=664
Score = 30.4 bits (67), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 23/76 (30%), Positives = 36/76 (47%), Gaps = 13/76 (17%)
Query 101 GMKKNRYGNIEDLVVDIK---VVT-TRGCMHACNSSPRISSGPSLQQLFLGSEGIYGIIT 156
G+K +R+ N+ D + D+K VVT R +I G L + GI+
Sbjct 418 GLKSDRFFNLRDTIADVKRNRVVTGLRFVKQNRIFHLQIQEGELLPR---------GIVN 468
Query 157 QVLLKIKPIPEYKVYD 172
Q L+ KP+ +Y V+D
Sbjct 469 QSTLEWKPVEKYNVFD 484
> At3g45090
Length=716
Score = 30.0 bits (66), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 21/63 (33%), Positives = 32/63 (50%), Gaps = 7/63 (11%)
Query 82 EPDSMEFSTVG--GWVATRASGMKKNRYGNIEDLVVDIKVVTTRGCMHACNSSPRISSGP 139
EP +++F G W + A+ G+++DL DI +R +C SSPR S GP
Sbjct 491 EPVNLQFGHFGISAWPSDGATS----HAGSVDDLRADIIETGSRH-YSSCCSSPRTSDGP 545
Query 140 SLQ 142
S +
Sbjct 546 SKE 548
> At1g48950
Length=608
Score = 29.6 bits (65), Expect = 3.3, Method: Composition-based stats.
Identities = 12/25 (48%), Positives = 17/25 (68%), Gaps = 0/25 (0%)
Query 74 PYGVTLGHEPDSMEFSTVGGWVATR 98
P G TL H+P S++F+ GG AT+
Sbjct 392 PKGGTLQHQPSSLKFTIAGGPPATK 416
> At3g63440
Length=504
Score = 29.6 bits (65), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 29/121 (23%), Positives = 51/121 (42%), Gaps = 5/121 (4%)
Query 46 KILYLDRDALLMWVEAGAVGASIEERLRPYGVTLGHEPDSMEFSTVGGWVATRASGMKKN 105
++ +D A + V G + +I YG+ D + TVGG ++ +
Sbjct 121 QVYSVDSPAPYVDVSGGELWINILHETLKYGLAPKSWTDYLHL-TVGGTLSNAGISGQAF 179
Query 106 RYGNIEDLVVDIKVVTTRGCMHACNSSPRISSGPSLQQLFLGSEGIYGIITQVLLKIKPI 165
R+G V +++VT +G + C L LG G +GIIT+ + ++P
Sbjct 180 RHGPQISNVHQLEIVTGKGEILNCTKR----QNSDLFNGVLGGLGQFGIITRARIALEPA 235
Query 166 P 166
P
Sbjct 236 P 236
Lambda K H
0.322 0.139 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3022542264
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40