bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0734_orf1
Length=140
Score E
Sequences producing significant alignments: (Bits) Value
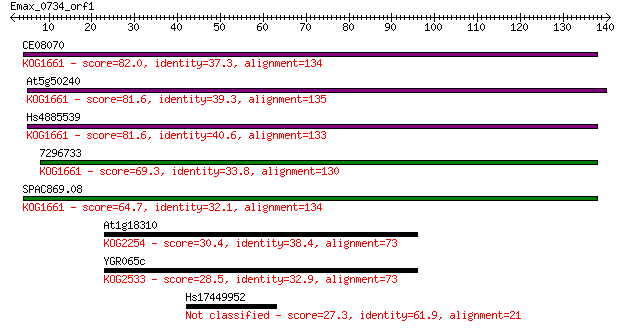
CE08070 82.0 3e-16
At5g50240 81.6 4e-16
Hs4885539 81.6 5e-16
7296733 69.3 2e-12
SPAC869.08 64.7 5e-11
At1g18310 30.4 1.1
YGR065c 28.5 4.2
Hs17449952 27.3 8.5
> CE08070
Length=225
Score = 82.0 bits (201), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 50/134 (37%), Positives = 70/134 (52%), Gaps = 20/134 (14%)
Query 4 GGIAIGIDYIQGLVDLSKKNVTAGFPHLMQSRNFALLKGDGWAGGPSWGAPYDAIHVGAA 63
G +GI+++ LV+LS+KN+ ++ N +++GDG G APY+AIHVGAA
Sbjct 103 NGTVVGIEHMPQLVELSEKNIRKHHSEQLERGNVIIIEGDGRQGFAE-KAPYNAIHVGAA 161
Query 64 AAAVPAALLQQLAIGGKMVIPVECHEGLITLEGLGDRQIDGDCDGGQALVVISKNKEGAV 123
+ VP AL QLA GG+M+IPVE DG Q + I K G +
Sbjct 162 SKGVPKALTDQLAEGGRMMIPVE------------------QVDGNQVFMQIDK-INGKI 202
Query 124 SVKYITSVVYVPLV 137
K + V+YVPL
Sbjct 203 EQKIVEHVIYVPLT 216
> At5g50240
Length=269
Score = 81.6 bits (200), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 53/136 (38%), Positives = 73/136 (53%), Gaps = 22/136 (16%)
Query 5 GIAIGIDYIQGLVDLSKKNVTAGFP-HLMQSRNFALLKGDGWAGGPSWGAPYDAIHVGAA 63
G +G+D+I LVD+S KN+ ++ + +L GDG G + APYDAIHVGAA
Sbjct 148 GRVVGVDHIPELVDMSIKNIEKSVAASFLKKGSLSLHVGDGRKGWQEF-APYDAIHVGAA 206
Query 64 AAAVPAALLQQLAIGGKMVIPVECHEGLITLEGLGDRQIDGDCDGGQALVVISKNKEGAV 123
A+ +P LL QL GG+MVIP+ + Q L VI KN++G++
Sbjct 207 ASEIPQPLLDQLKPGGRMVIPLGTY--------------------FQELKVIDKNEDGSI 246
Query 124 SVKYITSVVYVPLVRQ 139
V TSV YVPL +
Sbjct 247 KVHTETSVRYVPLTSR 262
> Hs4885539
Length=227
Score = 81.6 bits (200), Expect = 5e-16, Method: Compositional matrix adjust.
Identities = 54/133 (40%), Positives = 70/133 (52%), Gaps = 19/133 (14%)
Query 5 GIAIGIDYIQGLVDLSKKNVTAGFPHLMQSRNFALLKGDGWAGGPSWGAPYDAIHVGAAA 64
G IGID+I+ LVD S NV P L+ S L+ GDG G APYDAIHVGAAA
Sbjct 104 GKVIGIDHIKELVDDSVNNVRKDDPTLLSSGRVQLVVGDGRMGYAE-EAPYDAIHVGAAA 162
Query 65 AAVPAALLQQLAIGGKMVIPVECHEGLITLEGLGDRQIDGDCDGGQALVVISKNKEGAVS 124
VP AL+ QL GG++++PV G G Q L K ++G++
Sbjct 163 PVVPQALIDQLKPGGRLILPV------------------GPAGGNQMLEQYDKLQDGSIK 204
Query 125 VKYITSVVYVPLV 137
+K + V+YVPL
Sbjct 205 MKPLMGVIYVPLT 217
> 7296733
Length=226
Score = 69.3 bits (168), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 44/130 (33%), Positives = 64/130 (49%), Gaps = 19/130 (14%)
Query 8 IGIDYIQGLVDLSKKNVTAGFPHLMQSRNFALLKGDGWAGGPSWGAPYDAIHVGAAAAAV 67
+GI++ LV SK N+ ++ S +++GDG G P APY+AIHVGAAA
Sbjct 112 VGIEHQAELVRRSKANLNTDDRSMLDSGQLLIVEGDGRKGYPP-NAPYNAIHVGAAAPDT 170
Query 68 PAALLQQLAIGGKMVIPVECHEGLITLEGLGDRQIDGDCDGGQALVVISKNKEGAVSVKY 127
P L+ QLA GG++++PV G G Q + K+ G V +
Sbjct 171 PTELINQLASGGRLIVPV------------------GPDGGSQYMQQYDKDANGKVEMTR 212
Query 128 ITSVVYVPLV 137
+ V+YVPL
Sbjct 213 LMGVMYVPLT 222
> SPAC869.08
Length=230
Score = 64.7 bits (156), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 43/140 (30%), Positives = 67/140 (47%), Gaps = 27/140 (19%)
Query 4 GGIAIGIDYIQGLVDLSKKNVTAGFPH------LMQSRNFALLKGDGWAGGPSWGAPYDA 57
G GI++I LV+ SKKN+ H + + + + GDG G S +DA
Sbjct 103 NGTVKGIEHIPQLVETSKKNLLKDINHDEVLMEMYKEKRLQINVGDGRMGT-SEDEKFDA 161
Query 58 IHVGAAAAAVPAALLQQLAIGGKMVIPVECHEGLITLEGLGDRQIDGDCDGGQALVVISK 117
IHVGA+A+ +P L+ QL GK++IP+ + Q + +I K
Sbjct 162 IHVGASASELPQKLVDQLKSPGKILIPIGTY--------------------SQNIYLIEK 201
Query 118 NKEGAVSVKYITSVVYVPLV 137
N++G +S + + V YVPL
Sbjct 202 NEQGKISKRTLFPVRYVPLT 221
> At1g18310
Length=649
Score = 30.4 bits (67), Expect = 1.1, Method: Composition-based stats.
Identities = 28/75 (37%), Positives = 34/75 (45%), Gaps = 8/75 (10%)
Query 23 NVTAGFPHLMQSRNFALLKGDGWAGGPS--WGAPYDAIHVGAAAAAVPAALLQQLAIGGK 80
N + +P L RNF L K WAGG + W A A AALL LA G K
Sbjct 459 NSNSSYPRL---RNFDLFKLHSWAGGLTEFWDGRNQESTSEAVNAYYSAALL-GLAYGDK 514
Query 81 MVIPVECHEGLITLE 95
+ VE ++TLE
Sbjct 515 HL--VETASTIMTLE 527
> YGR065c
Length=593
Score = 28.5 bits (62), Expect = 4.2, Method: Composition-based stats.
Identities = 24/73 (32%), Positives = 35/73 (47%), Gaps = 11/73 (15%)
Query 23 NVTAGFPHLMQSRNFALLKGDGWAGGPSWGAPYDAIHVGAAAAAVPAALLQQLAIGGKMV 82
+VT+G L+QSR F L G G W DAI A ++P A++ I G
Sbjct 266 SVTSG---LLQSRIFKSLNGVHGLAGWRWMFLIDAI-----AISLPTAIIGFFVIPG--- 314
Query 83 IPVECHEGLITLE 95
+P +C+ +T E
Sbjct 315 VPSKCYSLFLTDE 327
> Hs17449952
Length=1205
Score = 27.3 bits (59), Expect = 8.5, Method: Composition-based stats.
Identities = 13/23 (56%), Positives = 16/23 (69%), Gaps = 4/23 (17%)
Query 42 GDGWAGGPSWGAPYD--AIHVGA 62
GDGWA GP APYD ++H G+
Sbjct 1062 GDGWAQGPP--APYDGPSVHAGS 1082
Lambda K H
0.318 0.139 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1571531578
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40