bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0739_orf1
Length=131
Score E
Sequences producing significant alignments: (Bits) Value
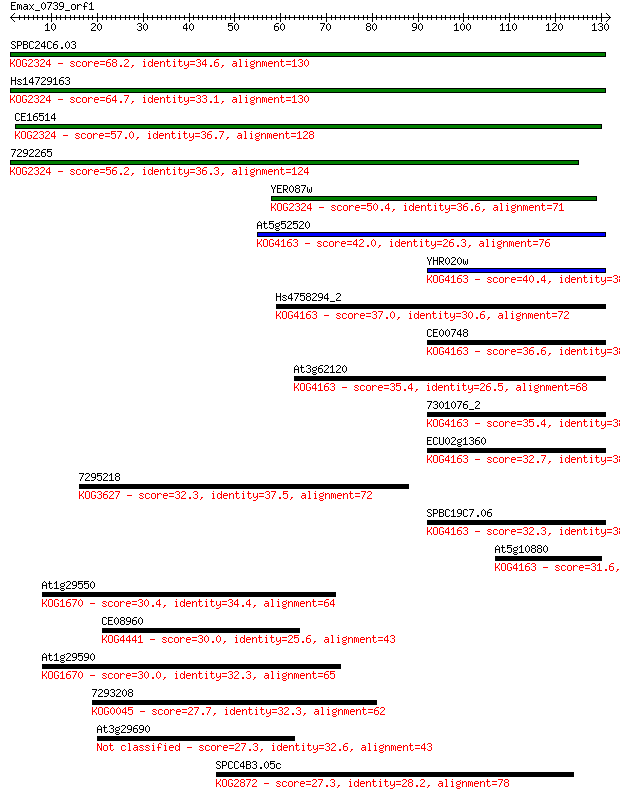
SPBC24C6.03 68.2 4e-12
Hs14729163 64.7 4e-11
CE16514 57.0 8e-09
7292265 56.2 2e-08
YER087w 50.4 9e-07
At5g52520 42.0 3e-04
YHR020w 40.4 0.001
Hs4758294_2 37.0 0.010
CE00748 36.6 0.012
At3g62120 35.4 0.025
7301076_2 35.4 0.028
ECU02g1360 32.7 0.16
7295218 32.3 0.21
SPBC19C7.06 32.3 0.23
At5g10880 31.6 0.37
At1g29550 30.4 0.94
CE08960 30.0 1.2
At1g29590 30.0 1.2
7293208 27.7 5.6
At3g29690 27.3 6.9
SPCC4B3.05c 27.3 8.0
> SPBC24C6.03
Length=425
Score = 68.2 bits (165), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 45/141 (31%), Positives = 69/141 (48%), Gaps = 12/141 (8%)
Query 1 PVYRFQADPGTMGGPCSHEYHVSSILGSDRIV----------SGVGPLNESKKDASAEDM 50
P +A G +GG SHE+H +G D I S + L+++ D S +
Sbjct 196 PFVMVKAATGNIGGNLSHEFHYRHPVGEDVIYTCPSCHYSTNSEMLDLSKTSSDISCPNC 255
Query 51 EYHQNSST-LEVGHCFQLPDTYCKAARTRFTDSRGALRVPLMNSYGLGVSRLLAHLALVH 109
S+T +EVGH F L Y + + V M YG+GVSRL+A +A V
Sbjct 256 NDQLTSTTAIEVGHAFYLGKIYSSKFNAT-VEVKNKQEVLHMGCYGIGVSRLIAAVAHVT 314
Query 110 QDSKGLLFPPQVAPFCAAILP 130
+D+KGL++P +AP+ ++P
Sbjct 315 KDAKGLVWPSSIAPWKVLVVP 335
> Hs14729163
Length=402
Score = 64.7 bits (156), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 43/141 (30%), Positives = 66/141 (46%), Gaps = 15/141 (10%)
Query 1 PVYRFQADPGTMGGPCSHEYHVSSILGSDRIVSGVGPLNESKKDASAEDMEYHQ------ 54
P + QAD GT+GG SHE+ + +G DR+ + P A+ E ++ Q
Sbjct 167 PFVKVQADVGTIGGTVSHEFQLPVDIGEDRL--AICP--RCSFSANMETLDLSQMNCPAC 222
Query 55 -----NSSTLEVGHCFQLPDTYCKAARTRFTDSRGALRVPLMNSYGLGVSRLLAHLALVH 109
+ +EVGH F L Y +FT+ G + M YGLGV+R+LA V
Sbjct 223 QGPLTKTKGIEVGHTFYLGTKYSSIFNAQFTNVCGKPTLAEMGCYGLGVTRILAAAIEVL 282
Query 110 QDSKGLLFPPQVAPFCAAILP 130
+ +P +AP+ A ++P
Sbjct 283 STEDCVRWPSLLAPYQACLIP 303
> CE16514
Length=454
Score = 57.0 bits (136), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 47/142 (33%), Positives = 67/142 (47%), Gaps = 23/142 (16%)
Query 2 VYRFQADPGTMGGPCSHEYHVSSILGSDRI-----------VSGVGPLNESKKDASAED- 49
V + +AD G GG SHEYH+ S L D + G GP +K + AE+
Sbjct 196 VIKVEADSGVHGGHISHEYHLKSSLEEDFVNFCENCKNHNKFEGPGP---AKCEKCAENS 252
Query 50 MEYHQNSSTLEVGHCFQLPDTYCKAARTRFTDSRGALRVPL-MNSYGLGVSRLL-AHLAL 107
+ Q ++E+ H F L Y +A +F PL M +G+GV+RLL A + L
Sbjct 253 SKTVQKIPSVEIAHTFHLGTKYSEALGAKFQGK------PLDMCCFGIGVTRLLPAAIDL 306
Query 108 VHQDSKGLLFPPQVAPFCAAIL 129
+ K L P +APF A I+
Sbjct 307 LSVSDKALRLPRAIAPFDAVII 328
> 7292265
Length=458
Score = 56.2 bits (134), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 45/135 (33%), Positives = 61/135 (45%), Gaps = 15/135 (11%)
Query 1 PVYRFQADPGTMGGPCSHEYHVSSILGSDRIV--SGVGPLNES---KKDASAEDMEYHQN 55
P + A G MGG SHEYH S +G D ++ S G S K AS N
Sbjct 199 PFVKVNAATGIMGGSVSHEYHYVSPVGEDNLLQCSSCGFAGNSEVVKAPASCPSC----N 254
Query 56 SSTL------EVGHCFQLPDTYCKAARTRFTDSRGALRVPLMNSYGLGVSRLLAHLALVH 109
SS L EV H F L D Y K F ++ G + +M YG+G++R++A V
Sbjct 255 SSDLKEVRGVEVAHTFLLGDKYSKPLGATFLNTTGKPQSLVMGCYGIGITRVIAAALEVL 314
Query 110 QDSKGLLFPPQVAPF 124
L +P +AP+
Sbjct 315 SSDHELRWPKLLAPY 329
> YER087w
Length=576
Score = 50.4 bits (119), Expect = 9e-07, Method: Composition-based stats.
Identities = 26/72 (36%), Positives = 40/72 (55%), Gaps = 1/72 (1%)
Query 58 TLEVGHCFQLPDTYCKAARTRFTDSRGALRVPL-MNSYGLGVSRLLAHLALVHQDSKGLL 116
++EVGH F L + Y K +F D + M YG+GVSRL+ +A + +DS G
Sbjct 378 SIEVGHIFLLGNKYSKPLNVKFVDKENKNETFVHMGCYGIGVSRLVGAIAELGRDSNGFR 437
Query 117 FPPQVAPFCAAI 128
+P +AP+ +I
Sbjct 438 WPAIMAPYKVSI 449
> At5g52520
Length=543
Score = 42.0 bits (97), Expect = 3e-04, Method: Composition-based stats.
Identities = 20/76 (26%), Positives = 36/76 (47%), Gaps = 1/76 (1%)
Query 55 NSSTLEVGHCFQLPDTYCKAARTRFTDSRGALRVPLMNSYGLGVSRLLAHLALVHQDSKG 114
+ L+ G L + +A T+F D G + S+ + +R + + + H D G
Sbjct 279 DRKALQAGTSHNLGQNFSRAFGTQFADENGERQHVWQTSWAVS-TRFVGGIIMTHGDDTG 337
Query 115 LLFPPQVAPFCAAILP 130
L+ PP++AP I+P
Sbjct 338 LMLPPKIAPIQVVIVP 353
> YHR020w
Length=688
Score = 40.4 bits (93), Expect = 0.001, Method: Composition-based stats.
Identities = 15/39 (38%), Positives = 29/39 (74%), Gaps = 1/39 (2%)
Query 92 NSYGLGVSRLLAHLALVHQDSKGLLFPPQVAPFCAAILP 130
NS+GL +R++ + ++H D+KGL+ PP+V+ F + ++P
Sbjct 443 NSWGLS-TRVIGVMVMIHSDNKGLVIPPRVSQFQSVVIP 480
> Hs4758294_2
Length=499
Score = 37.0 bits (84), Expect = 0.010, Method: Composition-based stats.
Identities = 22/74 (29%), Positives = 36/74 (48%), Gaps = 3/74 (4%)
Query 59 LEVGHCFQLPDTYCKAARTRFTDSR--GALRVPLMNSYGLGVSRLLAHLALVHQDSKGLL 116
++ G L + K F D + G + NS+GL +R + + +VH D+ GL+
Sbjct 223 IQGGTSHHLGQNFSKMFEIVFEDPKIPGEKQFAYQNSWGL-TTRTIGVMTMVHGDNMGLV 281
Query 117 FPPQVAPFCAAILP 130
PP+VA I+P
Sbjct 282 LPPRVACVQVVIIP 295
> CE00748
Length=581
Score = 36.6 bits (83), Expect = 0.012, Method: Composition-based stats.
Identities = 15/39 (38%), Positives = 25/39 (64%), Gaps = 1/39 (2%)
Query 92 NSYGLGVSRLLAHLALVHQDSKGLLFPPQVAPFCAAILP 130
NS+GL +R + + ++H D KGL+ PP+VA ++P
Sbjct 346 NSWGLS-TRTIGAMVMIHGDDKGLVLPPRVAAVQVIVVP 383
> At3g62120
Length=530
Score = 35.4 bits (80), Expect = 0.025, Method: Composition-based stats.
Identities = 18/68 (26%), Positives = 31/68 (45%), Gaps = 3/68 (4%)
Query 63 HCFQLPDTYCKAARTRFTDSRGALRVPLMNSYGLGVSRLLAHLALVHQDSKGLLFPPQVA 122
HC L + K F + + + NS+ +R + + + H D KGL+ PP+VA
Sbjct 275 HC--LGQNFAKMFEINFENEKAETEMVWQNSWAYS-TRTIGVMIMTHGDDKGLVLPPKVA 331
Query 123 PFCAAILP 130
++P
Sbjct 332 SVQVVVIP 339
> 7301076_2
Length=586
Score = 35.4 bits (80), Expect = 0.028, Method: Composition-based stats.
Identities = 15/39 (38%), Positives = 26/39 (66%), Gaps = 1/39 (2%)
Query 92 NSYGLGVSRLLAHLALVHQDSKGLLFPPQVAPFCAAILP 130
NS+G+ +R + + +VH D++GL+ PP VA A ++P
Sbjct 343 NSWGI-TTRTIGVMIMVHADNQGLVLPPHVACIQAIVVP 380
> ECU02g1360
Length=520
Score = 32.7 bits (73), Expect = 0.16, Method: Composition-based stats.
Identities = 15/39 (38%), Positives = 25/39 (64%), Gaps = 1/39 (2%)
Query 92 NSYGLGVSRLLAHLALVHQDSKGLLFPPQVAPFCAAILP 130
NS+G+ +R + A++H D+ GL+ PP+VA I+P
Sbjct 282 NSWGI-TTRSIGIAAMIHSDNLGLVLPPRVAMTQVVIVP 319
> 7295218
Length=259
Score = 32.3 bits (72), Expect = 0.21, Method: Compositional matrix adjust.
Identities = 27/80 (33%), Positives = 37/80 (46%), Gaps = 10/80 (12%)
Query 16 CSHEYHVSSILGSDRIVSGVG-PLNESKKDASAEDMEYHQNSSTL-------EVGHCFQL 67
C H+ S++ +RIV GV P++ + AS + SS L GHC Q
Sbjct 13 CGHK--TSALSPQERIVGGVEVPIHLTPWLASITVHGNYSCSSALITSLWLVTAGHCVQY 70
Query 68 PDTYCKAARTRFTDSRGALR 87
PD+Y A + FTD G R
Sbjct 71 PDSYSVRAGSTFTDGGGQRR 90
> SPBC19C7.06
Length=716
Score = 32.3 bits (72), Expect = 0.23, Method: Composition-based stats.
Identities = 15/39 (38%), Positives = 23/39 (58%), Gaps = 1/39 (2%)
Query 92 NSYGLGVSRLLAHLALVHQDSKGLLFPPQVAPFCAAILP 130
NS+GL +R + +VH D KGL PP +A + ++P
Sbjct 468 NSWGLS-TRTIGVAVMVHGDDKGLKLPPAIALVQSVVVP 505
> At5g10880
Length=309
Score = 31.6 bits (70), Expect = 0.37, Method: Compositional matrix adjust.
Identities = 11/23 (47%), Positives = 16/23 (69%), Gaps = 0/23 (0%)
Query 107 LVHQDSKGLLFPPQVAPFCAAIL 129
+ H D KGL+FPP+VAP ++
Sbjct 89 MTHGDDKGLVFPPKVAPVQVVVI 111
> At1g29550
Length=240
Score = 30.4 bits (67), Expect = 0.94, Method: Compositional matrix adjust.
Identities = 22/66 (33%), Positives = 29/66 (43%), Gaps = 12/66 (18%)
Query 8 DPGTMGGPCSHEYHVSSILGSDRIVSGVGPLNESKKDASAEDMEYHQNSST--LEVGHCF 65
DP T P E HVS+I + +SG +K S E Y ST ++ HCF
Sbjct 18 DPNTTTSPSPKEKHVSAI----KAISG------DEKAPSKEKKNYASKKSTTVIQKSHCF 67
Query 66 QLPDTY 71
Q T+
Sbjct 68 QNSWTF 73
> CE08960
Length=430
Score = 30.0 bits (66), Expect = 1.2, Method: Composition-based stats.
Identities = 11/43 (25%), Positives = 25/43 (58%), Gaps = 0/43 (0%)
Query 21 HVSSILGSDRIVSGVGPLNESKKDASAEDMEYHQNSSTLEVGH 63
H I+G +++ + + +ES +++ A + +YH +S + E H
Sbjct 39 HRQGIIGVEKMTNHIFSDSESDRESGASEFDYHSDSQSEETQH 81
> At1g29590
Length=285
Score = 30.0 bits (66), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 21/67 (31%), Positives = 31/67 (46%), Gaps = 12/67 (17%)
Query 8 DPGTMGGPCSHEYHVSSILGSDRIVSG--VGPLNESKKDASAEDMEYHQNSSTLEVGHCF 65
DP T P E HVS+I + +SG P E K AS ++++ ++ HCF
Sbjct 63 DPNTTTSPSPIEKHVSAI----KAISGDEKAPSKEKKNYAS------KKSTTVIQKSHCF 112
Query 66 QLPDTYC 72
Q T+
Sbjct 113 QNSWTFW 119
> 7293208
Length=1043
Score = 27.7 bits (60), Expect = 5.6, Method: Composition-based stats.
Identities = 20/62 (32%), Positives = 26/62 (41%), Gaps = 5/62 (8%)
Query 19 EYHVSSILGSDRIVSGVGPLNESKKDASAEDMEYHQNSSTLEVGHCFQLPDTYCKAARTR 78
EY +S L D + V LN + + YH + LE GH +P TY A
Sbjct 775 EYMLSECLQKDFFKNHVSYLNSDY--GNTRHVSYHTH---LEAGHYVLIPTTYEPAEEAH 829
Query 79 FT 80
FT
Sbjct 830 FT 831
> At3g29690
Length=180
Score = 27.3 bits (59), Expect = 6.9, Method: Compositional matrix adjust.
Identities = 14/48 (29%), Positives = 25/48 (52%), Gaps = 5/48 (10%)
Query 20 YHVSSILGSDRIVSGVGPLNESKKDASAEDME-----YHQNSSTLEVG 62
Y ++ LG D V+GV L++S K + +E Y + + ++VG
Sbjct 64 YEATTFLGEDGFVNGVEILSDSVKGLGSRSLESVWEVYEEGTKNMKVG 111
> SPCC4B3.05c
Length=370
Score = 27.3 bits (59), Expect = 8.0, Method: Compositional matrix adjust.
Identities = 22/79 (27%), Positives = 36/79 (45%), Gaps = 2/79 (2%)
Query 46 SAEDMEYHQNSSTLEVGHCFQLPDTYCKAARTRFTDSRGALRVPLMNSYGLGVSRLLAHL 105
+ E +EYH+ + Q P+T C+ T +G L ++ S L + + L +
Sbjct 42 TNETIEYHKLRAKQSFFEMCQTPETACELTLQPVTRFKGLLDAAIIFSDILVIPQALG-M 100
Query 106 ALVHQDSKGLLFP-PQVAP 123
+V + KG FP P V P
Sbjct 101 QVVMLEQKGPHFPKPLVVP 119
Lambda K H
0.320 0.135 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1283105810
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40