bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0761_orf1
Length=103
Score E
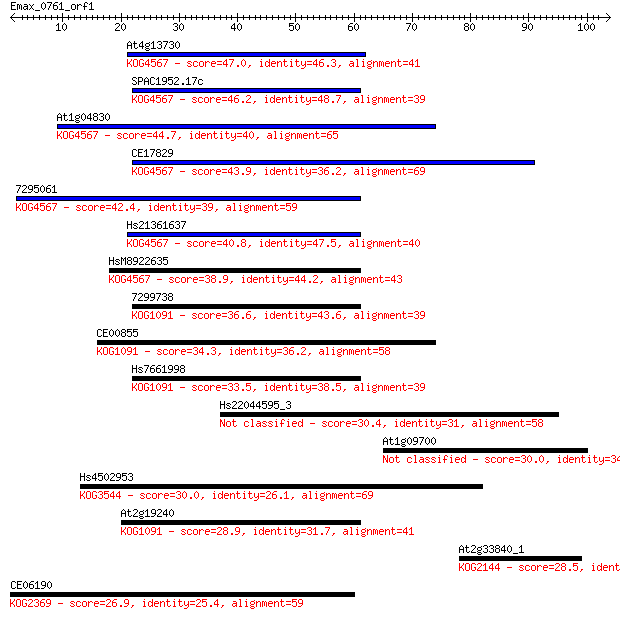
Sequences producing significant alignments: (Bits) Value
At4g13730 47.0 9e-06
SPAC1952.17c 46.2 1e-05
At1g04830 44.7 4e-05
CE17829 43.9 7e-05
7295061 42.4 2e-04
Hs21361637 40.8 6e-04
HsM8922635 38.9 0.002
7299738 36.6 0.012
CE00855 34.3 0.063
Hs7661998 33.5 0.095
Hs22044595_3 30.4 0.93
At1g09700 30.0 1.0
Hs4502953 30.0 1.1
At2g19240 28.9 2.7
At2g33840_1 28.5 3.4
CE06190 26.9 8.6
> At4g13730
Length=449
Score = 47.0 bits (110), Expect = 9e-06, Method: Composition-based stats.
Identities = 19/41 (46%), Positives = 27/41 (65%), Gaps = 0/41 (0%)
Query 21 KDHPLCRSATSEWRGYWVDAEVFDQINKDVFRTRPELCFFA 61
+DHPL TS W ++ D EV +QI +DV RT P++ FF+
Sbjct 206 EDHPLSLGTTSLWNNFFKDTEVLEQIERDVMRTHPDMHFFS 246
> SPAC1952.17c
Length=541
Score = 46.2 bits (108), Expect = 1e-05, Method: Composition-based stats.
Identities = 19/39 (48%), Positives = 28/39 (71%), Gaps = 0/39 (0%)
Query 22 DHPLCRSATSEWRGYWVDAEVFDQINKDVFRTRPELCFF 60
DHPL S S+W+ Y+ D ++ +QI+KD+ RT P+L FF
Sbjct 17 DHPLNTSDDSKWKEYFDDNQILEQIDKDIRRTLPDLSFF 55
> At1g04830
Length=451
Score = 44.7 bits (104), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 26/69 (37%), Positives = 38/69 (55%), Gaps = 6/69 (8%)
Query 9 LQSDSSIPLAAVK----DHPLCRSATSEWRGYWVDAEVFDQINKDVFRTRPELCFFALNP 64
L+S+S LA + DHPL S W Y+ D E +QI++DV RT P++ FF+
Sbjct 189 LKSESRCMLARSRITDEDHPLSLGKASIWNTYFQDTETIEQIDRDVKRTHPDIPFFS--G 246
Query 65 QQTIARQQQ 73
+ + AR Q
Sbjct 247 ESSFARSNQ 255
> CE17829
Length=484
Score = 43.9 bits (102), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 25/69 (36%), Positives = 37/69 (53%), Gaps = 5/69 (7%)
Query 22 DHPLCRSATSEWRGYWVDAEVFDQINKDVFRTRPELCFFALNPQQTIARQQQLHLYPPPP 81
DHPL TS+W+ ++ D +V QI+KDV R PE+ FF Q ++R H P
Sbjct 97 DHPLSDHPTSDWQAFFQDNKVLSQIDKDVRRLYPEIQFF-----QLLSRFPHQHGMKYPL 151
Query 82 TRHTQSQQQ 90
+R + Q+
Sbjct 152 SRRVINHQE 160
> 7295061
Length=403
Score = 42.4 bits (98), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 23/59 (38%), Positives = 31/59 (52%), Gaps = 2/59 (3%)
Query 2 STDGGGRLQSDSSIPLAAVKDHPLCRSATSEWRGYWVDAEVFDQINKDVFRTRPELCFF 60
S DGG + DS ++DHPL S W ++ D E QI+KDV R P++ FF
Sbjct 90 SVDGGDGDKVDSGG--VGLQDHPLSEGPESAWNTFFNDNEFLLQIDKDVRRLCPDISFF 146
> Hs21361637
Length=400
Score = 40.8 bits (94), Expect = 6e-04, Method: Composition-based stats.
Identities = 19/40 (47%), Positives = 25/40 (62%), Gaps = 0/40 (0%)
Query 21 KDHPLCRSATSEWRGYWVDAEVFDQINKDVFRTRPELCFF 60
+DHPL + S W Y+ D EV QI+KDV R P++ FF
Sbjct 98 EDHPLNPNPDSRWNTYFKDNEVLLQIDKDVRRLCPDISFF 137
> HsM8922635
Length=275
Score = 38.9 bits (89), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 19/43 (44%), Positives = 25/43 (58%), Gaps = 0/43 (0%)
Query 18 AAVKDHPLCRSATSEWRGYWVDAEVFDQINKDVFRTRPELCFF 60
+DHPL + S W Y+ D EV QI+KDV R P++ FF
Sbjct 95 VTFEDHPLNPNPDSRWNTYFKDNEVLLQIDKDVRRLCPDISFF 137
> 7299738
Length=654
Score = 36.6 bits (83), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 17/39 (43%), Positives = 23/39 (58%), Gaps = 0/39 (0%)
Query 22 DHPLCRSATSEWRGYWVDAEVFDQINKDVFRTRPELCFF 60
D PL +S S W Y+ D ++F I +DV RT P + FF
Sbjct 126 DDPLSQSTQSVWNQYFSDQDLFAVIRQDVVRTFPGVDFF 164
> CE00855
Length=585
Score = 34.3 bits (77), Expect = 0.063, Method: Compositional matrix adjust.
Identities = 21/58 (36%), Positives = 30/58 (51%), Gaps = 12/58 (20%)
Query 16 PLAAVKDHPLCRSATSEWRGYWVDAEVFDQINKDVFRTRPELCFFALNPQQTIARQQQ 73
PLA+++ +P W ++ D ++ D I KDV RT PE+ FF Q T RQ
Sbjct 100 PLASIEQNP--------WNTFFEDNDLRDIIGKDVSRTFPEIEFF----QNTSIRQMM 145
> Hs7661998
Length=795
Score = 33.5 bits (75), Expect = 0.095, Method: Compositional matrix adjust.
Identities = 15/39 (38%), Positives = 23/39 (58%), Gaps = 0/39 (0%)
Query 22 DHPLCRSATSEWRGYWVDAEVFDQINKDVFRTRPELCFF 60
++PL + S W ++ D E+ I +DV RT PE+ FF
Sbjct 139 NNPLSQDEGSLWNKFFQDKELRSMIEQDVKRTFPEMQFF 177
> Hs22044595_3
Length=214
Score = 30.4 bits (67), Expect = 0.93, Method: Compositional matrix adjust.
Identities = 18/59 (30%), Positives = 26/59 (44%), Gaps = 1/59 (1%)
Query 37 WVDAEVFDQINKDVFRTRPELCFFALNPQQTIARQ-QQLHLYPPPPTRHTQSQQQQQNE 94
W A D + +FR ++L+P + AR Q LHL P P + H + Q E
Sbjct 96 WPLARTADTPGRGLFRAHLPRPRWSLSPARGQARSGQALHLLPAPASLHPEGLPAQGGE 154
> At1g09700
Length=441
Score = 30.0 bits (66), Expect = 1.0, Method: Composition-based stats.
Identities = 12/35 (34%), Positives = 21/35 (60%), Gaps = 0/35 (0%)
Query 65 QQTIARQQQLHLYPPPPTRHTQSQQQQQNEHVPRL 99
++T+A+ + + PP PT H Q+ Q ++ E P L
Sbjct 243 KRTVAKNPEDIIIPPQPTDHCQNDQSEKIETTPNL 277
> Hs4502953
Length=1690
Score = 30.0 bits (66), Expect = 1.1, Method: Composition-based stats.
Identities = 18/72 (25%), Positives = 28/72 (38%), Gaps = 3/72 (4%)
Query 13 SSIPLAAVKDHPLCRSATSEWRGYWVDAEV---FDQINKDVFRTRPELCFFALNPQQTIA 69
S++P A H +C A R YW+ + ++++ R C P Q +A
Sbjct 1518 STLPFAYCNIHQVCHYAQRNDRSYWLASAAPLPMMPLSEEAIRPYVSRCAVCEAPAQAVA 1577
Query 70 RQQQLHLYPPPP 81
Q PP P
Sbjct 1578 VHSQDQSIPPCP 1589
> At2g19240
Length=840
Score = 28.9 bits (63), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 13/42 (30%), Positives = 26/42 (61%), Gaps = 1/42 (2%)
Query 20 VKDHPLCRSATSEWRGYWVDAEVFDQINKDVFRTRPE-LCFF 60
+ D+PL ++ S W ++ +AE+ +++D+ R PE C+F
Sbjct 81 IIDNPLSQNPNSTWGQFFRNAELEKTLDQDLSRLYPEHWCYF 122
> At2g33840_1
Length=379
Score = 28.5 bits (62), Expect = 3.4, Method: Composition-based stats.
Identities = 9/21 (42%), Positives = 14/21 (66%), Gaps = 0/21 (0%)
Query 78 PPPPTRHTQSQQQQQNEHVPR 98
PPPP+ T++ Q Q +H P+
Sbjct 358 PPPPSSSTRTSQNQPPQHTPQ 378
> CE06190
Length=417
Score = 26.9 bits (58), Expect = 8.6, Method: Composition-based stats.
Identities = 15/59 (25%), Positives = 31/59 (52%), Gaps = 2/59 (3%)
Query 1 VSTDGGGRLQSDSSIPLAAVKDHPLCRSATSEWRGYWVDAEVFDQINKDVFRTRPELCF 59
V DGG +L+S+ + + V H +C T+++ W++ ++F + D + +L F
Sbjct 39 VPGDGGSQLESNLTGKPSVV--HYVCSKQTADYFDLWLNLQLFTPLVIDCWADNMQLVF 95
Lambda K H
0.318 0.133 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1177719780
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40