bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0781_orf1
Length=159
Score E
Sequences producing significant alignments: (Bits) Value
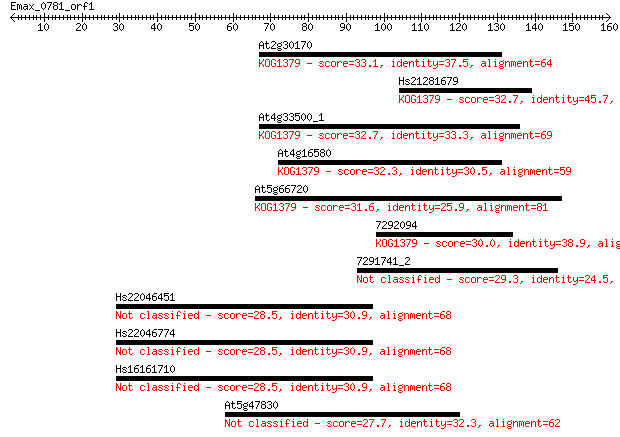
At2g30170 33.1 0.22
Hs21281679 32.7 0.33
At4g33500_1 32.7 0.34
At4g16580 32.3 0.40
At5g66720 31.6 0.76
7292094 30.0 2.1
7291741_2 29.3 3.4
Hs22046451 28.5 5.6
Hs22046774 28.5 6.2
Hs16161710 28.5 6.2
At5g47830 27.7 9.6
> At2g30170
Length=283
Score = 33.1 bits (74), Expect = 0.22, Method: Compositional matrix adjust.
Identities = 24/68 (35%), Positives = 34/68 (50%), Gaps = 6/68 (8%)
Query 67 ETPPLQP-LQFVVAGKSIQHPQKAALKSTNADAFAADQ---NVIGIADGVSTVEDEGLDP 122
E PL+P L V +I HP K ++ DAF V+ +ADGVS ++ +DP
Sbjct 37 EIQPLRPELSLSVGIHAIPHPDK--VEKGGEDAFFVSSYRGGVMAVADGVSGWAEQDVDP 94
Query 123 SELPAELL 130
S EL+
Sbjct 95 SLFSKELM 102
> Hs21281679
Length=304
Score = 32.7 bits (73), Expect = 0.33, Method: Compositional matrix adjust.
Identities = 16/38 (42%), Positives = 23/38 (60%), Gaps = 3/38 (7%)
Query 104 NVIGIADGVSTVEDEGLDPSELPAELLGLC---LKECR 138
+V+G+ADGV D G+DPS+ L+ C +KE R
Sbjct 72 DVLGVADGVGGWRDYGVDPSQFSGTLMRTCERLVKEGR 109
> At4g33500_1
Length=702
Score = 32.7 bits (73), Expect = 0.34, Method: Composition-based stats.
Identities = 23/69 (33%), Positives = 32/69 (46%), Gaps = 4/69 (5%)
Query 67 ETPPLQPLQFVVAGKSIQHPQKAALKSTNADAFAADQNVIGIADGVSTVEDEGLDPSELP 126
ET L+P++ S ++ AL F + N IGIADGVS EG++
Sbjct 458 ETASLEPIK----AASGRNNDVQALAGREDAYFISHHNWIGIADGVSQWSFEGINKGMYA 513
Query 127 AELLGLCLK 135
EL+ C K
Sbjct 514 QELMSNCEK 522
> At4g16580
Length=335
Score = 32.3 bits (72), Expect = 0.40, Method: Compositional matrix adjust.
Identities = 18/59 (30%), Positives = 29/59 (49%), Gaps = 0/59 (0%)
Query 72 QPLQFVVAGKSIQHPQKAALKSTNADAFAADQNVIGIADGVSTVEDEGLDPSELPAELL 130
+PL+ V + HP K A +A A++ +G+ADGV + G+D EL+
Sbjct 82 KPLKLVSGSCYLPHPDKEATGGEDAHFICAEEQALGVADGVGGWAELGIDAGYYSRELM 140
> At5g66720
Length=414
Score = 31.6 bits (70), Expect = 0.76, Method: Compositional matrix adjust.
Identities = 21/86 (24%), Positives = 39/86 (45%), Gaps = 5/86 (5%)
Query 66 RETPP-----LQPLQFVVAGKSIQHPQKAALKSTNADAFAADQNVIGIADGVSTVEDEGL 120
+E+PP L+ L+ V + HP+K A +A ++ IG+ADGV + G+
Sbjct 155 QESPPTTTTSLKSLRLVSGSCYLPHPEKEATGGEDAHFICDEEQAIGVADGVGGWAEVGV 214
Query 121 DPSELPAELLGLCLKECRARSLNSVV 146
+ EL+ + + + S +
Sbjct 215 NAGLFSRELMSYSVSAIQEQHKGSSI 240
> 7292094
Length=321
Score = 30.0 bits (66), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 14/36 (38%), Positives = 20/36 (55%), Gaps = 0/36 (0%)
Query 98 AFAADQNVIGIADGVSTVEDEGLDPSELPAELLGLC 133
A A +V+G+ADGV G+DP E + L+ C
Sbjct 85 ASTASADVMGVADGVGGWRSYGIDPGEFSSFLMRTC 120
> 7291741_2
Length=945
Score = 29.3 bits (64), Expect = 3.4, Method: Composition-based stats.
Identities = 13/53 (24%), Positives = 27/53 (50%), Gaps = 0/53 (0%)
Query 93 STNADAFAADQNVIGIADGVSTVEDEGLDPSELPAELLGLCLKECRARSLNSV 145
S N + D+N++G+ D DPSE+ +++L A+++N++
Sbjct 642 SVNLSKWEIDENILGLEDSSKKAAGASDDPSEITSDVLRKAENAIFAKAINAI 694
> Hs22046451
Length=990
Score = 28.5 bits (62), Expect = 5.6, Method: Compositional matrix adjust.
Identities = 21/70 (30%), Positives = 31/70 (44%), Gaps = 2/70 (2%)
Query 29 PLYCLPFFRFSLLWCCR--GMSCCACECGEGREAESFVMRETPPLQPLQFVVAGKSIQHP 86
P+ C + S L + G S CE G G E E + PP+ L+ V K+ H
Sbjct 465 PIQCFKLEKVSSLSLTQLAGPSSATCESGAGSEVEVDMFLRKPPMASLRKQVLTKASDHM 524
Query 87 QKAALKSTNA 96
++ L S+ A
Sbjct 525 PESLLASSPA 534
> Hs22046774
Length=994
Score = 28.5 bits (62), Expect = 6.2, Method: Compositional matrix adjust.
Identities = 21/70 (30%), Positives = 31/70 (44%), Gaps = 2/70 (2%)
Query 29 PLYCLPFFRFSLLWCCR--GMSCCACECGEGREAESFVMRETPPLQPLQFVVAGKSIQHP 86
P+ C + S L + G S CE G G E E + PP+ L+ V K+ H
Sbjct 469 PIQCFKLEKVSSLSLTQLAGPSSATCESGAGSEVEVDMFLRKPPMASLRKQVLTKASDHM 528
Query 87 QKAALKSTNA 96
++ L S+ A
Sbjct 529 PESLLASSPA 538
> Hs16161710
Length=994
Score = 28.5 bits (62), Expect = 6.2, Method: Compositional matrix adjust.
Identities = 21/70 (30%), Positives = 31/70 (44%), Gaps = 2/70 (2%)
Query 29 PLYCLPFFRFSLLWCCR--GMSCCACECGEGREAESFVMRETPPLQPLQFVVAGKSIQHP 86
P+ C + S L + G S CE G G E E + PP+ L+ V K+ H
Sbjct 469 PIQCFKLEKVSSLSLTQLAGPSSATCESGAGSEVEVDMFLRKPPMASLRKQVLTKASDHM 528
Query 87 QKAALKSTNA 96
++ L S+ A
Sbjct 529 PESLLASSPA 538
> At5g47830
Length=202
Score = 27.7 bits (60), Expect = 9.6, Method: Compositional matrix adjust.
Identities = 20/64 (31%), Positives = 29/64 (45%), Gaps = 2/64 (3%)
Query 58 REAESFVMRETPPLQPLQFVVAGKSIQHPQKAALK--STNADAFAADQNVIGIADGVSTV 115
RE ++ET + L + A +I Q AL +T+ D F Q V A G+S +
Sbjct 99 RELYKRRIQETASMDQLASIFAECAITEAQPLALDEPTTSKDLFGTKQTVTADAHGISIL 158
Query 116 EDEG 119
E G
Sbjct 159 EKSG 162
Lambda K H
0.322 0.136 0.441
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2136300674
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40