bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0791_orf1
Length=154
Score E
Sequences producing significant alignments: (Bits) Value
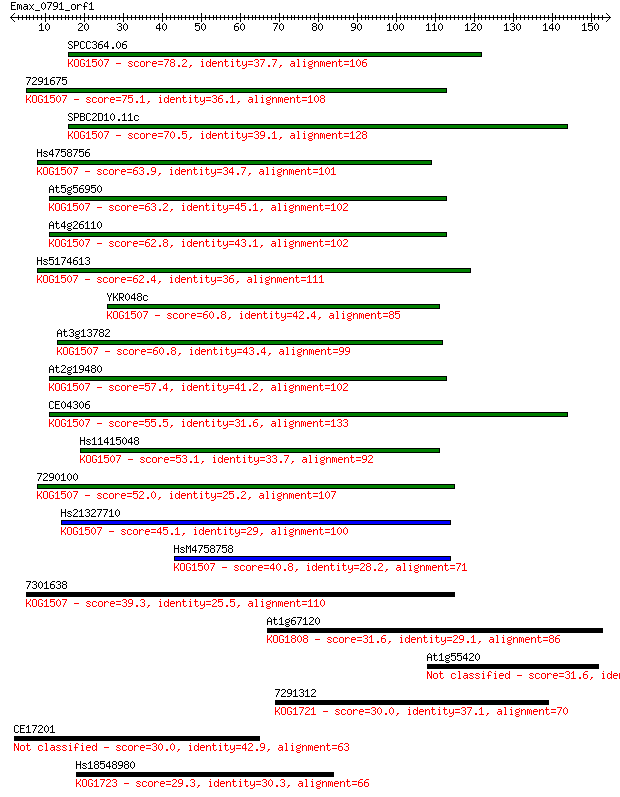
SPCC364.06 78.2 6e-15
7291675 75.1 5e-14
SPBC2D10.11c 70.5 1e-12
Hs4758756 63.9 1e-10
At5g56950 63.2 2e-10
At4g26110 62.8 3e-10
Hs5174613 62.4 3e-10
YKR048c 60.8 9e-10
At3g13782 60.8 9e-10
At2g19480 57.4 9e-09
CE04306 55.5 5e-08
Hs11415048 53.1 2e-07
7290100 52.0 5e-07
Hs21327710 45.1 5e-05
HsM4758758 40.8 0.001
7301638 39.3 0.003
At1g67120 31.6 0.59
At1g55420 31.6 0.64
7291312 30.0 2.0
CE17201 30.0 2.0
Hs18548980 29.3 2.8
> SPCC364.06
Length=393
Score = 78.2 bits (191), Expect = 6e-15, Method: Compositional matrix adjust.
Identities = 40/106 (37%), Positives = 64/106 (60%), Gaps = 4/106 (3%)
Query 16 SNKIEWYKEKDITKKIIIRKQRNKKTKQIRTITETVNNNSFFNLFNCHDIPSDDKLQQMD 75
+K++W + D+T + + +KQRNK TKQ R + +V +SFFN FN P ++ +
Sbjct 246 GDKVDWKENADLTVRTVTKKQRNKNTKQTRVVKVSVPRDSFFNFFN----PPTPPSEEDE 301
Query 76 EKEVEELQMVVEADYDIGITIRDKIIPRAVEWYLGQVDDDSDFDDY 121
E E EL ++E DY IG ++K+IPRAVEW+ G+ ++D +
Sbjct 302 ESESPELDELLELDYQIGEDFKEKLIPRAVEWFTGEALALENYDGF 347
> 7291675
Length=370
Score = 75.1 bits (183), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 39/109 (35%), Positives = 66/109 (60%), Gaps = 8/109 (7%)
Query 5 AAGAAELIKCISNKIEWYKEKDITKKIIIRKQRNKKTKQIRTITETVNNNSFFNLFNCHD 64
A E+ KC I W K+ ++T K I +KQ++K+ +RTI + V +SFFN F+ +
Sbjct 222 AFEGPEIYKCTGCTINWEKKMNLTVKTIRKKQKHKERGAVRTIVKQVPTDSFFNFFSPPE 281
Query 65 IPSDDKLQQMDEKEV-EELQMVVEADYDIGITIRDKIIPRAVEWYLGQV 112
+PS D++EV ++ Q ++ D++IG +R +IIP+AV +Y G +
Sbjct 282 VPS-------DQEEVDDDSQQILATDFEIGHFLRARIIPKAVLYYTGDI 323
> SPBC2D10.11c
Length=379
Score = 70.5 bits (171), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 50/131 (38%), Positives = 80/131 (61%), Gaps = 13/131 (9%)
Query 16 SNKIEWYK-EKDITKKIIIRKQRNKKTKQIRTITETVNNNSFFNLFNCHDIPSDDKLQQM 74
+KI W K EK++T ++ +KQRN+KT Q R + TV N+SFFN F+ + D+ +
Sbjct 250 GDKINWIKPEKNLTVRVETKKQRNRKTNQTRLVRTTVPNDSFFNFFSPPQLDDDESDDGL 309
Query 75 DEKEVEELQMVVEADYDIGITIRDKIIPRAVEWYLGQVDDDSDFDDYDINDEEFDSEE-- 132
D+K ++E DY +G +D+IIP A++ +L ++ D D++ DEE DSE+
Sbjct 310 DDK-----TELLELDYQLGEVFKDQIIPLAIDCFL----EEGDLSDFNQMDEE-DSEDAY 359
Query 133 SEEEEDSSDKE 143
++EE+ SSD E
Sbjct 360 TDEEDLSSDDE 370
> Hs4758756
Length=391
Score = 63.9 bits (154), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 35/101 (34%), Positives = 64/101 (63%), Gaps = 7/101 (6%)
Query 8 AAELIKCISNKIEWYKEKDITKKIIIRKQRNKKTKQIRTITETVNNNSFFNLFNCHDIPS 67
E++ C +I+W K K++T K I +KQ++K +RT+T+TV+N+SFFN F ++P
Sbjct 249 GPEIMGCTGCQIDWKKGKNVTLKTIKKKQKHKGRGTVRTVTKTVSNDSFFNFFAPPEVPE 308
Query 68 DDKLQQMDEKEVEELQMVVEADYDIGITIRDKIIPRAVEWY 108
L ++ + ++ AD++IG +R++IIPR+V ++
Sbjct 309 SGDLD-------DDAEAILAADFEIGHFLRERIIPRSVLYF 342
> At5g56950
Length=368
Score = 63.2 bits (152), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 46/102 (45%), Positives = 65/102 (63%), Gaps = 3/102 (2%)
Query 11 LIKCISNKIEWYKEKDITKKIIIRKQRNKKTKQIRTITETVNNNSFFNLFNCHDIPSDDK 70
L K I +I+WY K +T+KI+ +K + + IT+T + SFFN FN +P DD
Sbjct 200 LEKAIGTEIDWYPGKCLTQKILKKKPKKGAKNA-KPITKTEDCESFFNFFNPPQVPDDD- 257
Query 71 LQQMDEKEVEELQMVVEADYDIGITIRDKIIPRAVEWYLGQV 112
+ +DE+ EELQ ++E DYDIG TIR+KIIP AV W+ G+
Sbjct 258 -EDIDEERAEELQNLMEQDYDIGSTIREKIIPHAVSWFTGEA 298
> At4g26110
Length=372
Score = 62.8 bits (151), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 44/102 (43%), Positives = 69/102 (67%), Gaps = 3/102 (2%)
Query 11 LIKCISNKIEWYKEKDITKKIIIRKQRNKKTKQIRTITETVNNNSFFNLFNCHDIPSDDK 70
L K + +I+WY K +T+KI+ +K+ K +K + IT+ + SFFN F+ ++P +D
Sbjct 201 LEKAMGTEIDWYPGKCLTQKIL-KKKPKKGSKNTKPITKLEDCESFFNFFSPPEVPDED- 258
Query 71 LQQMDEKEVEELQMVVEADYDIGITIRDKIIPRAVEWYLGQV 112
+ +DE+ E+LQ ++E DYDIG TIR+KIIPRAV W+ G+
Sbjct 259 -EDIDEERAEDLQNLMEQDYDIGSTIREKIIPRAVSWFTGEA 299
> Hs5174613
Length=375
Score = 62.4 bits (150), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 40/112 (35%), Positives = 67/112 (59%), Gaps = 7/112 (6%)
Query 8 AAELIKCISNKIEWYKEKDITKKIIIRKQRNKKTKQIRTITETVNNNSFFNLFNCHDIPS 67
E++ C I+W K K++T K I +KQ++K +RTIT+ V N SFFN FN
Sbjct 241 GPEIVDCDGCTIDWKKGKNVTVKTIKKKQKHKGRGTVRTITKQVPNESFFNFFNPLKASG 300
Query 68 DDKLQQMDEKEVEELQMVVEADYDIGITIRDKIIPRAVEWYLGQ-VDDDSDF 118
D + +D E+ + + +D++IG R++I+PRAV ++ G+ ++DD +F
Sbjct 301 DG--ESLD----EDSEFTLASDFEIGHFFRERIVPRAVLYFTGEAIEDDDNF 346
> YKR048c
Length=417
Score = 60.8 bits (146), Expect = 9e-10, Method: Compositional matrix adjust.
Identities = 36/85 (42%), Positives = 56/85 (65%), Gaps = 4/85 (4%)
Query 26 DITKKIIIRKQRNKKTKQIRTITETVNNNSFFNLFNCHDIPSDDKLQQMDEKEVEELQMV 85
++T + +RKQRNK TKQ+RTI + SFFN F+ I ++D+ ++++E E L +
Sbjct 282 NVTVDLEMRKQRNKTTKQVRTIEKITPIESFFNFFDPPKIQNEDQDEELEEDLEERLAL- 340
Query 86 VEADYDIGITIRDKIIPRAVEWYLG 110
DY IG ++DK+IPRAV+W+ G
Sbjct 341 ---DYSIGEQLKDKLIPRAVDWFTG 362
> At3g13782
Length=329
Score = 60.8 bits (146), Expect = 9e-10, Method: Compositional matrix adjust.
Identities = 43/104 (41%), Positives = 65/104 (62%), Gaps = 5/104 (4%)
Query 13 KCISNKIEWYKEKDITKKIIIRKQRNKKTKQIRTI--TETVNNNSFFNLFNCHDIPSDDK 70
K I IEW+ K +T K++++K+ K K++ I T+T N SFFN F +IP D+
Sbjct 205 KVIGTDIEWFPGKCLTHKVVVKKKTKKGPKKVNNIPMTKTENCESFFNFFKPPEIPEIDE 264
Query 71 LQQMDEKEV---EELQMVVEADYDIGITIRDKIIPRAVEWYLGQ 111
+ D+ + EELQ +++ DYDI +TIRDK+IP AV W+ G+
Sbjct 265 VDDYDDFDTIMTEELQNLMDQDYDIAVTIRDKLIPHAVSWFTGE 308
> At2g19480
Length=379
Score = 57.4 bits (137), Expect = 9e-09, Method: Compositional matrix adjust.
Identities = 42/102 (41%), Positives = 62/102 (60%), Gaps = 3/102 (2%)
Query 11 LIKCISNKIEWYKEKDITKKIIIRKQRNKKTKQIRTITETVNNNSFFNLFNCHDIPSDDK 70
L K + +IEWY K +T+KI+ +K+ K +K + IT+T + SFFN F+ +P DD+
Sbjct 200 LEKALGTEIEWYPGKCLTQKIL-KKKPKKGSKNTKPITKTEDCESFFNFFSPPQVPDDDE 258
Query 71 LQQMDEKEVEELQMVVEADYDIGITIRDKIIPRAVEWYLGQV 112
D + + QM E DYDIG TI++KII AV W+ G+
Sbjct 259 DLDDDMADELQGQM--EHDYDIGSTIKEKIISHAVSWFTGEA 298
> CE04306
Length=316
Score = 55.5 bits (132), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 42/133 (31%), Positives = 73/133 (54%), Gaps = 13/133 (9%)
Query 11 LIKCISNKIEWYKEKDITKKIIIRKQRNKKTKQIRTITETVNNNSFFNLFNCHDIPSDDK 70
+I+ + + IEW ++ K ++K++ K + +T+TV +SFFN F P K
Sbjct 196 VIRAVGDTIEW-EDGKNVTKKAVKKKQKKGANAGKFLTKTVKADSFFNFFE----PPKSK 250
Query 71 LQQMDEKEVEELQMVVEADYDIGITIRDKIIPRAVEWYLGQVDDDSDFDDYDINDEEFDS 130
++ ++++ E+ + +E DY++G IRD IIPRAV +Y G++ D FD F
Sbjct 251 DERNEDEDDEQAEEFLELDYEMGQAIRDTIIPRAVLFYTGELQSDDMFD--------FPG 302
Query 131 EESEEEEDSSDKE 143
E+ ++ D SD E
Sbjct 303 EDGDDVSDFSDDE 315
> Hs11415048
Length=460
Score = 53.1 bits (126), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 31/92 (33%), Positives = 51/92 (55%), Gaps = 14/92 (15%)
Query 19 IEWYKEKDITKKIIIRKQRNKKTKQIRTITETVNNNSFFNLFNCHDIPSDDKLQQMDEKE 78
I+W + K++T K I +KQ+++ IRT+TE +SFFN F+ H I S+ + D +
Sbjct 333 IDWNEGKNVTLKTIKKKQKHRIWGTIRTVTEDFPKDSFFNFFSPHGITSNGR----DGND 388
Query 79 VEELQMVVEADYDIGITIRDKIIPRAVEWYLG 110
D+ +G +R IIPR+V ++ G
Sbjct 389 ----------DFLLGHNLRTYIIPRSVLFFSG 410
> 7290100
Length=362
Score = 52.0 bits (123), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 27/107 (25%), Positives = 54/107 (50%), Gaps = 11/107 (10%)
Query 8 AAELIKCISNKIEWYKEKDITKKIIIRKQRNKKTKQIRTITETVNNNSFFNLFNCHDIPS 67
E+++C I W ++T + + ++RN+ + +T+ + SFF F P
Sbjct 245 GPEIVRCEGCHIHWRDGSNLTLQTVESRRRNRAHR----VTKVMPRESFFRFFAP---PQ 297
Query 68 DDKLQQMDEKEVEELQMVVEADYDIGITIRDKIIPRAVEWYLGQVDD 114
L DEK ++++ D+++G +R +I+P+AV +Y G + D
Sbjct 298 ALDLSLADEKT----KLILGNDFEVGFLLRTQIVPKAVLFYTGDLVD 340
> Hs21327710
Length=506
Score = 45.1 bits (105), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 29/100 (29%), Positives = 52/100 (52%), Gaps = 9/100 (9%)
Query 14 CISNKIEWYKEKDITKKIIIRKQRNKKTKQIRTITETVNNNSFFNLFNCHDIPSDDKLQQ 73
C KI+W + KD+T + R T +I V N SFFN F+ +IP KL+
Sbjct 399 CKGCKIDWRRGKDVTVTT--TQSRTTATGEIEIQPRVVPNASFFNFFSPPEIPMIGKLEP 456
Query 74 MDEKEVEELQMVVEADYDIGITIRDKIIPRAVEWYLGQVD 113
++ +++ D++IG + D +I +++ +Y G+V+
Sbjct 457 RED-------AILDEDFEIGQILHDNVILKSIYYYTGEVN 489
> HsM4758758
Length=506
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 20/71 (28%), Positives = 39/71 (54%), Gaps = 7/71 (9%)
Query 43 QIRTITETVNNNSFFNLFNCHDIPSDDKLQQMDEKEVEELQMVVEADYDIGITIRDKIIP 102
+I V N SFFN F+ +IP KL+ ++ +++ D++IG + D +I
Sbjct 426 EIEIQPRVVPNASFFNFFSPPEIPMIGKLEPRED-------AILDEDFEIGQILHDNVIL 478
Query 103 RAVEWYLGQVD 113
+++ +Y G+V+
Sbjct 479 KSIYYYTGEVN 489
> 7301638
Length=272
Score = 39.3 bits (90), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 28/110 (25%), Positives = 52/110 (47%), Gaps = 24/110 (21%)
Query 5 AAGAAELIKCISNKIEWYKEKDITKKIIIRKQRNKKTKQIRTITETVNNNSFFNLFNCHD 64
A AE+ KC I+W + KD TK TE SFF F+
Sbjct 159 AYDGAEIYKCEGCVIDWKQTKDQTK------------------TEN-QEPSFFEFFSPPL 199
Query 65 IPSDDKLQQMDEKEVEELQMVVEADYDIGITIRDKIIPRAVEWYLGQVDD 114
+P D +D + + +++ D+++G +++++IP+AV ++ G++ D
Sbjct 200 LPED----TLDPNYCD-VNAMLQNDFEVGFYLKERVIPKAVIFFTGEIAD 244
> At1g67120
Length=5138
Score = 31.6 bits (70), Expect = 0.59, Method: Composition-based stats.
Identities = 25/86 (29%), Positives = 43/86 (50%), Gaps = 9/86 (10%)
Query 67 SDDKLQQMDEKEVEELQMVVEADYDIGITIRDKIIPRAVEWYLGQVDDDSDFDDYDINDE 126
+ DK ++ +++ VE+ D+D + +KI + E LG + D D + D +D
Sbjct 4437 TSDKPEEGNDENVEQ------DDFDDTDNLEEKI--QTKEEALGGLTPDVDNEQID-DDM 4487
Query 127 EFDSEESEEEEDSSDKEPPRSHKGHH 152
E D E E+ED++ +E P S H
Sbjct 4488 EMDKTEEVEKEDANQQEEPCSEDQKH 4513
> At1g55420
Length=725
Score = 31.6 bits (70), Expect = 0.64, Method: Composition-based stats.
Identities = 19/49 (38%), Positives = 25/49 (51%), Gaps = 5/49 (10%)
Query 108 YLGQVDDDSDFD---DYDINDEEFDSEESEEEED--SSDKEPPRSHKGH 151
YLG V DD D D+D+ E D E+S + D +SD + SH H
Sbjct 124 YLGDVSDDDGEDSHSDHDLGASEDDGEDSHSDHDLGASDDDGEDSHSDH 172
> 7291312
Length=835
Score = 30.0 bits (66), Expect = 2.0, Method: Composition-based stats.
Identities = 26/70 (37%), Positives = 34/70 (48%), Gaps = 12/70 (17%)
Query 69 DKLQQMDEKEVEELQMVVEADYDIGITIRDKIIPRAVEWYLGQVDDDSDFDDYDINDEEF 128
D+ Q MDE E EE QM E D D I ++I Q++DD DD D + +
Sbjct 405 DQQQAMDEIEDEEEQMKPEEDQDTFII--EEI----------QLEDDEMLDDPDGEEIDQ 452
Query 129 DSEESEEEED 138
D E EE+D
Sbjct 453 DCEYIGEEQD 462
> CE17201
Length=934
Score = 30.0 bits (66), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 27/65 (41%), Positives = 32/65 (49%), Gaps = 8/65 (12%)
Query 2 TGAAAGAAELIKCISNKIEWYKEKDITKKIIIRKQRNKKTKQIRTITETVN--NNSFFNL 59
T AA G EL K S+ KDI K I+ K+ +K K+ TITETV SF NL
Sbjct 398 TTAAEGKLELFKKSSD------FKDIYKLILPAKKAAQKLKEAPTITETVTKIQTSFGNL 451
Query 60 FNCHD 64
D
Sbjct 452 TTFVD 456
> Hs18548980
Length=190
Score = 29.3 bits (64), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 20/66 (30%), Positives = 37/66 (56%), Gaps = 6/66 (9%)
Query 18 KIEWYKEKDITKKIIIRKQRNKKTKQIRTITETVNNNSFFNLFNCHDIPSDDKLQQMDEK 77
++E K+K K II R ++NK+ ++++ I E N +P + K +Q++EK
Sbjct 111 RVEEIKQKRQAKCIINRLRKNKELQKVQDIQEVKQNIHLTR------VPLEGKEKQLEEK 164
Query 78 EVEELQ 83
V++LQ
Sbjct 165 TVQQLQ 170
Lambda K H
0.312 0.131 0.376
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1997677330
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40