bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0858_orf2
Length=265
Score E
Sequences producing significant alignments: (Bits) Value
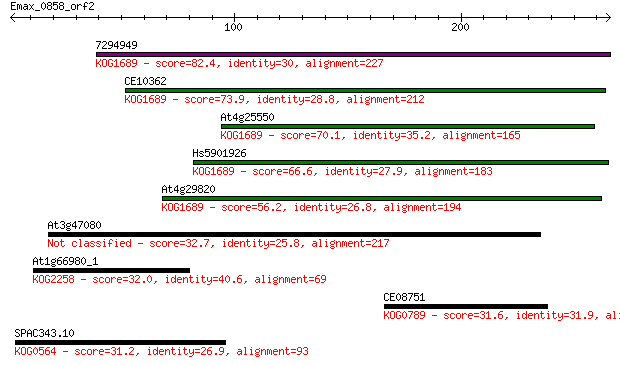
7294949 82.4 9e-16
CE10362 73.9 3e-13
At4g25550 70.1 4e-12
Hs5901926 66.6 5e-11
At4g29820 56.2 6e-08
At3g47080 32.7 0.89
At1g66980_1 32.0 1.2
CE08751 31.6 1.6
SPAC343.10 31.2 2.1
> 7294949
Length=237
Score = 82.4 bits (202), Expect = 9e-16, Method: Compositional matrix adjust.
Identities = 68/232 (29%), Positives = 101/232 (43%), Gaps = 32/232 (13%)
Query 39 LSSGFVECQRNGDFEREGLSASMDATSSIEHRQGNFKTISKTTSAPRDVELEWAVFPESN 98
++S V + + R G DA SS + G K ++ + R + L +P +N
Sbjct 1 MASSQVSNKSGSGWPRRGSQGQADAASS--NNNGTQKYTNQALTINRTINL----YPLTN 54
Query 99 YSFEADSTLGNKWGSGSEEELLKKRQEAYFREGTRRSVAAIFLVHRAEYPHVLLLLDQQQ 158
Y+F L K S ++ +E + R G RRSV + LVH PHVLLL
Sbjct 55 YTFGTKEPLFEK--DPSVPSRFQRMREEFDRIGMRRSVEGVLLVHEHGLPHVLLL----- 107
Query 159 KKHSLLMFKY-----KTWQKPREVLHAKLAEYLIRPEQCSKRKWVAQQLSNEGPDMEVGE 213
+ FK + E L L+E L R + K++W+ V +
Sbjct 108 -QLGTTFFKLPGGELNAGEDEVEGLKRLLSETLGR-QDGVKQEWI------------VED 153
Query 214 FLGEWWRGEFDDDLVPYLPPHVTRPKERVRVHQVQLPHRRSFRVPMGFCLTV 265
+G WWR F+ PY+PPH+T+PKE R+ VQL + F VP + L
Sbjct 154 TIGNWWRPNFEPPQYPYIPPHITKPKEHKRLFLVQLHEKALFAVPKNYKLVA 205
> CE10362
Length=227
Score = 73.9 bits (180), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 61/216 (28%), Positives = 94/216 (43%), Gaps = 37/216 (17%)
Query 52 FEREGLSASM-DATSSIEHRQGNFKTISKTTSAPRDVELEWAVFPESNYSFEADSTLGNK 110
ER +SAS+ +A ++ + + +TI+ V+P +NY+F K
Sbjct 8 IERTTISASVPEAPANFDEKPPFNRTIN--------------VYPLTNYTFGTKDAQAEK 53
Query 111 WGSGSEEELLKKRQEAYFREGTRRSVAAIFLVHRAEYPHVLLLLDQQQKKHSLLMFKYKT 170
S E K+ ++ Y G RRSV A+ +VH PH+LLL
Sbjct 54 --DKSVPERFKRMKDEYEVMGMRRSVEAVLIVHEHSLPHILLL-----------QIGTTF 100
Query 171 WQKPREVLHAKLAEYLIRPEQCSKRKWVAQQL---SNEGPDMEVGEFLGEWWRGEFDDDL 227
++ P L L E + + + L E + + + +G WWR FD
Sbjct 101 YKLPGGELE------LGEDEISGVTRLLNETLGRTDGETNEWTIEDEIGNWWRPNFDPPR 154
Query 228 VPYLPPHVTRPKERVRVHQVQLPHRRSFRVPMGFCL 263
PY+P HVT+PKE ++ VQLP + +F VP F L
Sbjct 155 YPYIPAHVTKPKEHTKLLLVQLPSKSTFCVPKNFKL 190
> At4g25550
Length=210
Score = 70.1 bits (170), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 58/185 (31%), Positives = 83/185 (44%), Gaps = 34/185 (18%)
Query 94 FPESNYSF-------EADSTLGNKWGSGSEEELLKKRQEAYFREGTRRSVAAIFLVHRAE 146
+P SNYSF E D+++ ++ L + + Y +EG R SV I LV
Sbjct 10 YPLSNYSFGTKEPKLEKDTSVADR---------LARMKINYMKEGMRTSVEGILLVQEHN 60
Query 147 YPHVLLLLDQQQKKHSLLMFKYKTWQKPREV---LHAKLAEYLIRPEQCS-KRKWVAQQL 202
+PH+LLL Q ++ KP E L Y++ E KRK ++
Sbjct 61 HPHILLL----QIGNTFCKLP-GGRLKPGENGIQLPPFWVYYVVSAEADGLKRKLTSKLG 115
Query 203 SNEG---PDMEVGEFLGEWWRGEFDDDLVPYLPPHVTRPK------ERVRVHQVQLPHRR 253
N PD VGE + WWR F+ + PY PPH+T+PK E R++ V L +
Sbjct 116 GNSAALVPDWTVGECVATWWRPNFETMMYPYCPPHITKPKVVKKHNECKRLYIVHLSEKE 175
Query 254 SFRVP 258
F VP
Sbjct 176 YFAVP 180
> Hs5901926
Length=227
Score = 66.6 bits (161), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 51/184 (27%), Positives = 81/184 (44%), Gaps = 18/184 (9%)
Query 82 SAPRDVELEWAVFPESNYSFEADSTLGNKWGSGSEEELLKKRQEAYFREGTRRSVAAIFL 141
+ P +E ++P +NY+F L K S ++ +E + + G RR+V + +
Sbjct 28 TKPLTLERTINLYPLTNYTFGTKEPLYEK--DSSVAARFQRMREEFDKIGMRRTVEGVLI 85
Query 142 VHRAEYPHVLLLLDQQQKKHSLLMFKYKTWQ-KPREVLHAKLAEYLIRPEQCSKRKWVAQ 200
VH PHVLLL + FK + P E L + E ++ V Q
Sbjct 86 VHEHRLPHVLLL------QLGTTFFKLPGGELNPGEDEVEGLKRLMT--EILGRQDGVLQ 137
Query 201 QLSNEGPDMEVGEFLGEWWRGEFDDDLVPYLPPHVTRPKERVRVHQVQLPHRRSFRVPMG 260
D + + +G WWR F+ PY+P H+T+PKE ++ VQL + F VP
Sbjct 138 -------DWVIDDCIGNWWRPNFEPPQYPYIPAHITKPKEHKKLFLVQLQEKALFAVPKN 190
Query 261 FCLT 264
+ L
Sbjct 191 YKLV 194
> At4g29820
Length=185
Score = 56.2 bits (134), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 52/196 (26%), Positives = 84/196 (42%), Gaps = 54/196 (27%)
Query 68 EHRQGNFKTISKTTSAPRDV--ELEWAVFPESNYSFEADSTLGNKWGSGSEEELLKKRQE 125
E R + + IS T+ DV +L ++P S+Y F G+K ++E++ R
Sbjct 4 EARALDMEEISDNTTRRNDVVHDLMVDLYPLSSYYF------GSKEALRVKDEIISDR-- 55
Query 126 AYFREGTRRSVAAIFLVHRAEYPHVLLLLDQQQKKHSLLMFKYKTWQKPREVLHAKLAEY 185
+PHVLLL Q ++S+ KL
Sbjct 56 ---------------------HPHVLLL----QYRNSIF----------------KLPGG 74
Query 186 LIRPEQCSKRKWVAQQLSNEGPDMEVGEFLGEWWRGEFDDDLVPYLPPHVTRPKERVRVH 245
+RP + V L++ ++ VGE +G WWR F+ + P+LPP++ PKE ++
Sbjct 75 RLRP---GESGLVCCFLASLCINIAVGECIGMWWRPNFETLMYPFLPPNIKHPKECTKLF 131
Query 246 QVQLPHRRSFRVPMGF 261
V+LP + F VP F
Sbjct 132 LVRLPVHQQFVVPKNF 147
> At3g47080
Length=515
Score = 32.7 bits (73), Expect = 0.89, Method: Compositional matrix adjust.
Identities = 56/246 (22%), Positives = 100/246 (40%), Gaps = 36/246 (14%)
Query 18 ATGGEGTADPALNVSVDTSQELSSGFVECQRNGDFEREGLSASMDA-TSSIEHRQGNFKT 76
A+ G ++D A +V + QEL+S F + E + S ++D S HR G+ K
Sbjct 103 ASSGFFSSDEAFSVKM---QELASQFRNAGDEEEEENKQKSEAVDNDNDSNNHRFGSLKL 159
Query 77 ISKTTSAPRDVELEWA-VFPESNYSFEADST--------LGNKWGSGSEEELLKKRQEAY 127
+ ++ +E WA + S+ +A+S + K +EE LKK
Sbjct 160 LQESVPGLASLEAPWAEMVNHSSIERKANSVDLPLSLRIIKRKL----QEEALKKASATT 215
Query 128 FREGTRRSVAAIFLVHR----AEYPHVLLLLDQQQKKHSLLM--FKYKTWQKPREVLHAK 181
+ R + +F++ A V +L +++ H+ L+ F+ Q P +++
Sbjct 216 YCSINRAFSSMVFMIEELHSFALQTRVGVLKQVKKEMHASLLWIFQRVFSQTPTLMVYVM 275
Query 182 --LAEYLIRPEQCSKRKWVAQQLSNEGPDM-----------EVGEFLGEWWRGEFDDDLV 228
LA Y + + + + N+GPD E + G W G + D V
Sbjct 276 ILLANYTVHSVASNLPIAASPPVVNKGPDQTQQRIDFSSLKESTKLDGSKWLGSINFDKV 335
Query 229 PYLPPH 234
+LP H
Sbjct 336 SHLPRH 341
> At1g66980_1
Length=764
Score = 32.0 bits (71), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 28/80 (35%), Positives = 39/80 (48%), Gaps = 15/80 (18%)
Query 11 RGGMKMQATGGEGTADPALNVSVDTSQELSSGFVECQRNGDFEREGL--------SASMD 62
+ G+K+ A+G D A N S D E S FV+ NG+F +G+ SAS+D
Sbjct 299 KAGLKLYASGFANDVDIAYNYSWDPVSEYLS-FVD---NGNFSVDGMLSDFPLTASASVD 354
Query 63 ATSSI---EHRQGNFKTISK 79
S I +Q +F ISK
Sbjct 355 CFSHIGRNATKQVDFLVISK 374
> CE08751
Length=497
Score = 31.6 bits (70), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 23/82 (28%), Positives = 38/82 (46%), Gaps = 20/82 (24%)
Query 166 FKYKTWQKPREVLHAKLAEYLIRPEQCSK--------RKW--VAQQLSNEGPDMEVGEFL 215
K K+ QK E +H K + R ++C+K KW VAQ+ + ++ V EF
Sbjct 166 LKGKSKQKSEEKVHQKAKDDAKRGDECNKFKISDEKLTKWKLVAQETLKDDANIAVNEF- 224
Query 216 GEWWRGEFDDDLVPYLPPHVTR 237
+ + YLPPH+++
Sbjct 225 ---------EKVSGYLPPHISK 237
> SPAC343.10
Length=641
Score = 31.2 bits (69), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 25/95 (26%), Positives = 39/95 (41%), Gaps = 6/95 (6%)
Query 3 VRGRVWLRRGGMKMQATGGEGTADPALNVSVDTSQELSSGFVECQRNGDFEREGLSASMD 62
+ G W G A G +ADP + FVEC NG ++ ++ D
Sbjct 477 LNGNSWWTVGSQP--AVNGAPSADPVFGWGPKGGRVFQKAFVECFVNGKDLKDFITKWHD 534
Query 63 ATSSIEHRQGNFKTISKTTSAPRD--VELEWAVFP 95
+ + GN K+ T+AP+D + W V+P
Sbjct 535 -NPQVTYYAGNNKS-EFLTNAPKDGASAVTWGVYP 567
Lambda K H
0.318 0.134 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5649423594
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40