bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0871_orf1
Length=172
Score E
Sequences producing significant alignments: (Bits) Value
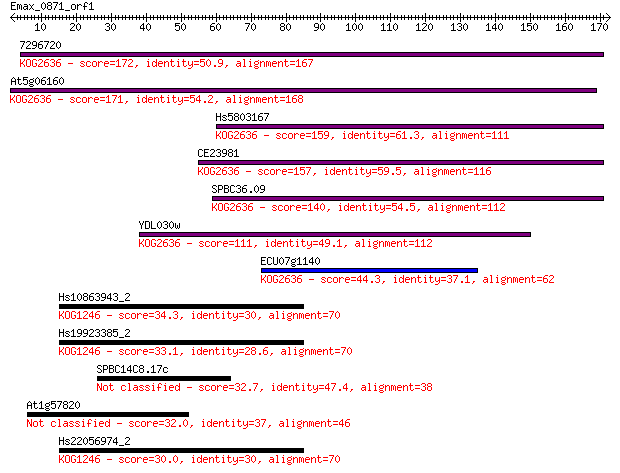
7296720 172 3e-43
At5g06160 171 6e-43
Hs5803167 159 2e-39
CE23981 157 7e-39
SPBC36.09 140 1e-33
YDL030w 111 8e-25
ECU07g1140 44.3 1e-04
Hs10863943_2 34.3 0.14
Hs19923385_2 33.1 0.30
SPBC14C8.17c 32.7 0.34
At1g57820 32.0 0.55
Hs22056974_2 30.0 2.2
> 7296720
Length=503
Score = 172 bits (436), Expect = 3e-43, Method: Compositional matrix adjust.
Identities = 85/167 (50%), Positives = 115/167 (68%), Gaps = 7/167 (4%)
Query 4 FVDLLQPQINQTIAHYQKKQSTNAEEALEEENDEDSNEEDAAAEEDSEAEEEEGPIYNPL 63
+ DLL Q T + Q+KQ+ E +DS+ E + ++ + + + ++ P YNP
Sbjct 331 YADLLSEQRAATKENVQRKQARTGGER------DDSDVEASESDNEDDPDADDVP-YNPK 383
Query 64 NLPLGFDGRPIPYWLYKLHGLGQEFKCEICGNFSYWGRRAFERHFSEWRHSFGMRCLKIP 123
NLPLG+DG+PIPYWLYKLHGL + CEICGNF+Y G +AF+RHF+EWRH+ GMRCL IP
Sbjct 384 NLPLGWDGKPIPYWLYKLHGLNISYNCEICGNFTYKGPKAFQRHFAEWRHAHGMRCLGIP 443
Query 124 NTTHFKEITKIEDAILLYEKLKQQAEGHAFKQDQELECEDAEGNVMS 170
NT HF +T+IEDAI L+EKLK Q + + DQE E ED+ GNV++
Sbjct 444 NTAHFANVTQIEDAITLWEKLKSQKQSERWVADQEEEFEDSLGNVVN 490
> At5g06160
Length=504
Score = 171 bits (434), Expect = 6e-43, Method: Compositional matrix adjust.
Identities = 91/168 (54%), Positives = 119/168 (70%), Gaps = 7/168 (4%)
Query 1 IKYFVDLLQPQINQTIAHYQKKQSTNAEEALEEENDEDSNEEDAAAEEDSEAEEEEGPIY 60
+K +LL I +T + KKQS EE E E++N E E+++E+G IY
Sbjct 329 VKKLCNLLDETIERTKQNIVKKQSLTYEEMEGEREGEEANTE-------LESDDEDGLIY 381
Query 61 NPLNLPLGFDGRPIPYWLYKLHGLGQEFKCEICGNFSYWGRRAFERHFSEWRHSFGMRCL 120
NPL LP+G+DG+PIPYWLYKLHGLGQEFKCEICGN+SYWGRRAFERHF EWRH GMRCL
Sbjct 382 NPLKLPIGWDGKPIPYWLYKLHGLGQEFKCEICGNYSYWGRRAFERHFKEWRHQHGMRCL 441
Query 121 KIPNTTHFKEITKIEDAILLYEKLKQQAEGHAFKQDQELECEDAEGNV 168
IPNT +F EIT IE+A L+++++++ + ++ + E E ED EGN+
Sbjct 442 GIPNTKNFNEITSIEEAKELWKRIQERQGVNKWRPELEEEYEDREGNI 489
> Hs5803167
Length=501
Score = 159 bits (403), Expect = 2e-39, Method: Compositional matrix adjust.
Identities = 68/111 (61%), Positives = 86/111 (77%), Gaps = 0/111 (0%)
Query 60 YNPLNLPLGFDGRPIPYWLYKLHGLGQEFKCEICGNFSYWGRRAFERHFSEWRHSFGMRC 119
YNP NLPLG+DG+PIPYWLYKLHGL + CEICGN++Y G +AF+RHF+EWRH+ GMRC
Sbjct 378 YNPKNLPLGWDGKPIPYWLYKLHGLNINYNCEICGNYTYRGPKAFQRHFAEWRHAHGMRC 437
Query 120 LKIPNTTHFKEITKIEDAILLYEKLKQQAEGHAFKQDQELECEDAEGNVMS 170
L IPNT HF +T+IEDA+ L+ KLK Q ++ D E E ED+ GNV++
Sbjct 438 LGIPNTAHFANVTQIEDAVSLWAKLKLQKASERWQPDTEEEYEDSSGNVVN 488
> CE23981
Length=500
Score = 157 bits (398), Expect = 7e-39, Method: Compositional matrix adjust.
Identities = 69/116 (59%), Positives = 88/116 (75%), Gaps = 0/116 (0%)
Query 55 EEGPIYNPLNLPLGFDGRPIPYWLYKLHGLGQEFKCEICGNFSYWGRRAFERHFSEWRHS 114
+E YNP NLPLG+DG+PIPYWLYKLHGL + CEICGN +Y G +AF++HF+EWRHS
Sbjct 372 DESAPYNPKNLPLGWDGKPIPYWLYKLHGLNLSYSCEICGNQTYKGPKAFQKHFNEWRHS 431
Query 115 FGMRCLKIPNTTHFKEITKIEDAILLYEKLKQQAEGHAFKQDQELECEDAEGNVMS 170
GMRCL IPNT+HF ITKI+DA+ L+ KLK + E + D + E ED+ GNV++
Sbjct 432 HGMRCLGIPNTSHFANITKIKDALDLWNKLKTEKEMAKWNPDIDEEYEDSSGNVVT 487
> SPBC36.09
Length=492
Score = 140 bits (353), Expect = 1e-33, Method: Compositional matrix adjust.
Identities = 61/112 (54%), Positives = 83/112 (74%), Gaps = 0/112 (0%)
Query 59 IYNPLNLPLGFDGRPIPYWLYKLHGLGQEFKCEICGNFSYWGRRAFERHFSEWRHSFGMR 118
IYNPL LPLG+DG+PIP+WL+KLHGLG+EF CEICGN+ Y GR+AF++HF+E RH +G++
Sbjct 368 IYNPLKLPLGWDGKPIPFWLWKLHGLGKEFPCEICGNYVYMGRKAFDKHFTEQRHIYGLK 427
Query 119 CLKIPNTTHFKEITKIEDAILLYEKLKQQAEGHAFKQDQELECEDAEGNVMS 170
CL I + F +IT I++A+ L++K K ++ E ED EGNVMS
Sbjct 428 CLGISPSPLFNQITSIDEALQLWQKYKVDSKKRETTMASLNEMEDDEGNVMS 479
> YDL030w
Length=530
Score = 111 bits (277), Expect = 8e-25, Method: Compositional matrix adjust.
Identities = 55/116 (47%), Positives = 78/116 (67%), Gaps = 4/116 (3%)
Query 38 DSNEEDAAAEEDSEAE----EEEGPIYNPLNLPLGFDGRPIPYWLYKLHGLGQEFKCEIC 93
DS E++ A + D E +EE ++PLG DG P+PYWLYKLHGL +E++CEIC
Sbjct 367 DSTEKEGAEQVDGEQRDGQLQEEHLSGKSFDMPLGPDGLPMPYWLYKLHGLDREYRCEIC 426
Query 94 GNFSYWGRRAFERHFSEWRHSFGMRCLKIPNTTHFKEITKIEDAILLYEKLKQQAE 149
N Y GRR FERHF+E RH + +RCL I ++ FK ITKI++A L++ ++ Q++
Sbjct 427 SNKVYNGRRTFERHFNEERHIYHLRCLGIEPSSVFKGITKIKEAQELWKNMQGQSQ 482
> ECU07g1140
Length=370
Score = 44.3 bits (103), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 23/62 (37%), Positives = 33/62 (53%), Gaps = 1/62 (1%)
Query 73 PIPYWLYKLHGLGQEFKCEICGNFSYWGRRAFERHFSEWRHSFGMRCLKIPNTTHFKEIT 132
+P WL + L F+CEICG +S +GR F+RHF E H+ G+ K IT
Sbjct 268 SVPRWLCRRKDLDIAFECEICG-YSGYGRDRFDRHFEEDSHARGVEMYGAKYCRVLKGIT 326
Query 133 KI 134
++
Sbjct 327 RV 328
> Hs10863943_2
Length=522
Score = 34.3 bits (77), Expect = 0.14, Method: Composition-based stats.
Identities = 21/74 (28%), Positives = 37/74 (50%), Gaps = 5/74 (6%)
Query 15 TIAHYQKKQSTNAEEALEEENDEDSNEEDAAAEE----DSEAEEEEGPIYNPLNLPLGFD 70
TIA Y + Q+++ +E+L EEN++ S+ +D + E D+ +GP + + D
Sbjct 152 TIAKYAQYQASSFQESLREENEKRSHHKDHSDSESTSSDNSGRRRKGP-FKTIKFGTNID 210
Query 71 GRPIPYWLYKLHGL 84
W +LH L
Sbjct 211 LSDDKKWKLQLHEL 224
> Hs19923385_2
Length=521
Score = 33.1 bits (74), Expect = 0.30, Method: Composition-based stats.
Identities = 20/74 (27%), Positives = 37/74 (50%), Gaps = 5/74 (6%)
Query 15 TIAHYQKKQSTNAEEALEEENDEDSNEEDAAAEEDSEAE----EEEGPIYNPLNLPLGFD 70
TIA Y + Q+++ +E+L EEN++ + +D + E + +E +GP + + D
Sbjct 152 TIAKYAQYQASSFQESLREENEKRTQHKDHSDNESTSSENSGRRRKGP-FKTIKFGTNID 210
Query 71 GRPIPYWLYKLHGL 84
W +LH L
Sbjct 211 LSDNKKWKLQLHEL 224
> SPBC14C8.17c
Length=526
Score = 32.7 bits (73), Expect = 0.34, Method: Composition-based stats.
Identities = 18/39 (46%), Positives = 23/39 (58%), Gaps = 1/39 (2%)
Query 26 NAEEALEEENDEDSNEEDAAAEEDSEAEE-EEGPIYNPL 63
+ EE +E + ED NE D E+ SE +E EE P NPL
Sbjct 56 SGEEDVEMDTMEDENENDEDTEQTSEKKETEETPKENPL 94
> At1g57820
Length=641
Score = 32.0 bits (71), Expect = 0.55, Method: Composition-based stats.
Identities = 17/47 (36%), Positives = 26/47 (55%), Gaps = 1/47 (2%)
Query 6 DLLQ-PQINQTIAHYQKKQSTNAEEALEEENDEDSNEEDAAAEEDSE 51
D LQ PQ+N+ +A +K T E+ E E++++ EEDSE
Sbjct 576 DFLQNPQVNREVAEVIEKLKTQEEDTAELEDEDEGECSGTTPEEDSE 622
> Hs22056974_2
Length=304
Score = 30.0 bits (66), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 21/74 (28%), Positives = 37/74 (50%), Gaps = 5/74 (6%)
Query 15 TIAHYQKKQSTNAEEALEEENDEDSNEEDAAAEE----DSEAEEEEGPIYNPLNLPLGFD 70
TIA Y + Q+++ +E+L EEN++ S+ +D + E D+ +GP + + D
Sbjct 152 TIAKYAQYQASSFQESLREENEKRSHHKDHSDSESTSSDNSGRRRKGP-FKTIKFGTNID 210
Query 71 GRPIPYWLYKLHGL 84
W +LH L
Sbjct 211 LSDDKKWKLQLHEL 224
Lambda K H
0.315 0.134 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2562785186
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40