bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
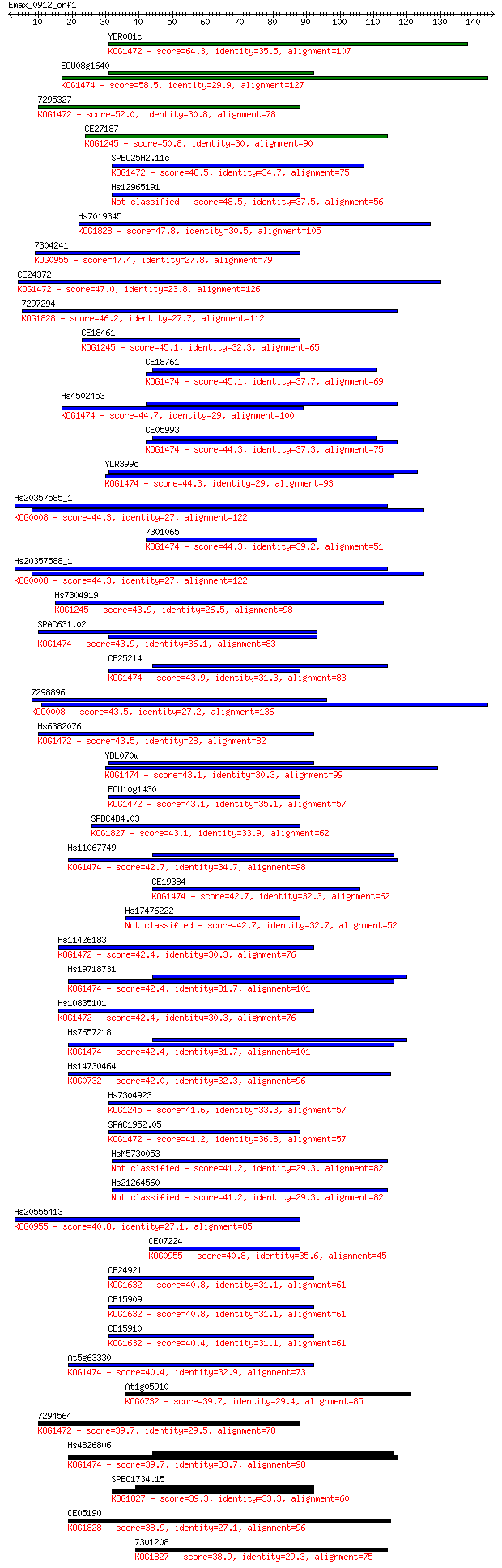
Query= Emax_0912_orf1
Length=145
Score E
Sequences producing significant alignments: (Bits) Value
YBR081c 64.3 8e-11
ECU08g1640 58.5 4e-09
7295327 52.0 4e-07
CE27187 50.8 9e-07
SPBC25H2.11c 48.5 4e-06
Hs12965191 48.5 4e-06
Hs7019345 47.8 8e-06
7304241 47.4 8e-06
CE24372 47.0 1e-05
7297294 46.2 2e-05
CE18461 45.1 4e-05
CE18761 45.1 5e-05
Hs4502453 44.7 5e-05
CE05993 44.3 8e-05
YLR399c 44.3 9e-05
Hs20357585_1 44.3 9e-05
7301065 44.3 9e-05
Hs20357588_1 44.3 9e-05
Hs7304919 43.9 9e-05
SPAC631.02 43.9 1e-04
CE25214 43.9 1e-04
7298896 43.5 1e-04
Hs6382076 43.5 1e-04
YDL070w 43.1 2e-04
ECU10g1430 43.1 2e-04
SPBC4B4.03 43.1 2e-04
Hs11067749 42.7 2e-04
CE19384 42.7 2e-04
Hs17476222 42.7 3e-04
Hs11426183 42.4 3e-04
Hs19718731 42.4 3e-04
Hs10835101 42.4 3e-04
Hs7657218 42.4 3e-04
Hs14730464 42.0 4e-04
Hs7304923 41.6 6e-04
SPAC1952.05 41.2 7e-04
HsM5730053 41.2 7e-04
Hs21264560 41.2 7e-04
Hs20555413 40.8 8e-04
CE07224 40.8 9e-04
CE24921 40.8 9e-04
CE15909 40.8 0.001
CE15910 40.4 0.001
At5g63330 40.4 0.001
At1g05910 39.7 0.002
7294564 39.7 0.002
Hs4826806 39.7 0.002
SPBC1734.15 39.3 0.003
CE05190 38.9 0.003
7301208 38.9 0.003
> YBR081c
Length=1332
Score = 64.3 bits (155), Expect = 8e-11, Method: Composition-based stats.
Identities = 38/108 (35%), Positives = 58/108 (53%), Gaps = 3/108 (2%)
Query 31 PFVNRVDDRVAPGYSVVVQHPMFLRKVLSKCRERQYTSVAAFKDDINLIVSNCLLFNPVG 90
PF+N+V R AP Y +++ M L VL K + QY S F DDI LI NCL +N
Sbjct 465 PFLNKVSKREAPNYHQIIKKSMDLNTVLKKLKSFQYDSKQEFVDDIMLIWKNCLTYN--S 522
Query 91 SPSAWLRDRAVQLQQLAMQRLAEQPQIA-EYEAALQQNAIHVQQQQQY 137
PS +LR A+ +Q+ ++Q + P I A L++ +++ + Y
Sbjct 523 DPSHFLRGHAIAMQKKSLQLIRMIPNITIRNRADLEKEIEDMEKDKDY 570
> ECU08g1640
Length=370
Score = 58.5 bits (140), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 27/61 (44%), Positives = 37/61 (60%), Gaps = 0/61 (0%)
Query 31 PFVNRVDDRVAPGYSVVVQHPMFLRKVLSKCRERQYTSVAAFKDDINLIVSNCLLFNPVG 90
PF+ VD + PGY V++ PM L+ + SK +R+Y SV F D+ LIV NC FN G
Sbjct 175 PFLEPVDGDLVPGYYSVIKEPMDLQTMRSKLEQRRYQSVEEFGRDLELIVENCKKFNAPG 234
Query 91 S 91
+
Sbjct 235 T 235
Score = 47.0 bits (110), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 31/130 (23%), Positives = 56/130 (43%), Gaps = 6/130 (4%)
Query 17 ARVVNLLLQQQTFKPFVNRVD--DRVAPGYSVVVQHPMFLRKVLSKCRERQYTSVAAFKD 74
++++ L + PF+ VD P Y ++HPM L + K ++Y F
Sbjct 21 SQILTRLKRNSNAPPFLEPVDPVKLGIPDYPEKIKHPMDLSTIRKKLDSKEYEGPEGFDG 80
Query 75 DINLIVSNCLLFNPVGSPSAWLRDRAVQLQQLAMQRLAEQPQ-IAEYEAALQQNAIHVQQ 133
D+ L+ SNC +NP P + + L+ + +A PQ I + + A+ +
Sbjct 81 DMRLMFSNCYTYNP---PGTVVHEMGKGLEAVYTDLMAGMPQEIPKKRKKTEMPAVGRPK 137
Query 134 QQQYSTYGYD 143
Q + + G D
Sbjct 138 QVKRNVKGSD 147
> 7295327
Length=3080
Score = 52.0 bits (123), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 24/78 (30%), Positives = 44/78 (56%), Gaps = 0/78 (0%)
Query 10 DALNVSFARVVNLLLQQQTFKPFVNRVDDRVAPGYSVVVQHPMFLRKVLSKCRERQYTSV 69
+ L + +V+ + + PFV+ V++ +AP Y +++ PM L K+ K +Y
Sbjct 748 EVLQIGMHKVLVYVKNHRDAWPFVDPVEEDIAPRYYSIIRRPMDLLKMEDKLDSGEYHKF 807
Query 70 AAFKDDINLIVSNCLLFN 87
+ F++D LIV+NC L+N
Sbjct 808 SEFRNDFRLIVNNCRLYN 825
> CE27187
Length=1427
Score = 50.8 bits (120), Expect = 9e-07, Method: Composition-based stats.
Identities = 27/90 (30%), Positives = 46/90 (51%), Gaps = 3/90 (3%)
Query 24 LQQQTFKPFVNRVDDRVAPGYSVVVQHPMFLRKVLSKCRERQYTSVAAFKDDINLIVSNC 83
++Q+ PF+ VD + P Y V++ PM LR +++K ++R Y ++D LI+SNC
Sbjct 1337 MRQECSWPFLQPVDSKEVPDYYDVIKRPMNLRTMMNKIKQRIYNKPIEVRNDFQLILSNC 1396
Query 84 LLFNPVGSPSAWLRDRAVQLQQLAMQRLAE 113
+N P + + +L RL E
Sbjct 1397 ETYN---EPENEIYKLSRELHDFMADRLDE 1423
> SPBC25H2.11c
Length=979
Score = 48.5 bits (114), Expect = 4e-06, Method: Composition-based stats.
Identities = 26/80 (32%), Positives = 43/80 (53%), Gaps = 5/80 (6%)
Query 32 FVNRVDDRVAPGYSVVVQHPMFLRKVLSKCRERQYTSVAAFKDDINLIVSNCLLFN---- 87
F+ +V R AP Y V++ PM L +L + Y S F D+ LI SNC L+N
Sbjct 324 FLTKVSKRDAPDYYTVIKEPMDLGTILRNLKNLHYNSKKEFVHDLMLIWSNCFLYNSHPD 383
Query 88 -PVGSPSAWLRDRAVQLQQL 106
P+ + +++D++++L L
Sbjct 384 HPLRVHAQFMKDKSLELINL 403
> Hs12965191
Length=233
Score = 48.5 bits (114), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 21/56 (37%), Positives = 31/56 (55%), Gaps = 0/56 (0%)
Query 32 FVNRVDDRVAPGYSVVVQHPMFLRKVLSKCRERQYTSVAAFKDDINLIVSNCLLFN 87
F V D +APGYS++++HPM + K +Y SV FK D L+ N + +N
Sbjct 45 FAFPVTDAIAPGYSMIIKHPMDFGTMKDKIVANEYKSVTEFKADFKLMCDNAMTYN 100
> Hs7019345
Length=652
Score = 47.8 bits (112), Expect = 8e-06, Method: Composition-based stats.
Identities = 32/113 (28%), Positives = 55/113 (48%), Gaps = 11/113 (9%)
Query 22 LLLQQQTFKP---FVNRVDDRVAPGYSVVVQHPMFLRKVLSKCRERQYTSVAAFKDDINL 78
L+ Q Q P F V D +APGYS++++HPM + K + Y S+ KD+ L
Sbjct 143 LMRQLQRKDPSAFFSFPVTDFIAPGYSMIIKHPMDFSTMKEKIKNNDYQSIEELKDNFKL 202
Query 79 IVSNCLLFNPVGSPSAWLRDRAVQL-----QQLAMQRLAEQPQIAEYEAALQQ 126
+ +N +++N P A +L + L+ +R+ Q ++ A LQ+
Sbjct 203 MCTNAMIYN---KPETIYYKAAKKLLHSGMKILSQERIQSLKQSIDFMADLQK 252
> 7304241
Length=1430
Score = 47.4 bits (111), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 22/79 (27%), Positives = 44/79 (55%), Gaps = 0/79 (0%)
Query 9 IDALNVSFARVVNLLLQQQTFKPFVNRVDDRVAPGYSVVVQHPMFLRKVLSKCRERQYTS 68
++ L + ++++ L + + + F VD P Y+ +V+ PM L + +K +E QY S
Sbjct 613 LNPLEAALNKLLDALEARDSMQIFREPVDTSEVPDYTDIVKQPMDLGTMRAKLKECQYNS 672
Query 69 VAAFKDDINLIVSNCLLFN 87
+ + D +L++ NCL +N
Sbjct 673 LEQLEADFDLMIQNCLAYN 691
> CE24372
Length=767
Score = 47.0 bits (110), Expect = 1e-05, Method: Composition-based stats.
Identities = 30/127 (23%), Positives = 61/127 (48%), Gaps = 4/127 (3%)
Query 4 HMQQPIDALNVSFARVVNLLLQQQTFKPFVNRVDDRVAPGYSVVVQHPMFLRKVLSKCRE 63
H+ + D+L+ ++ L + PF + VD + P Y ++HP+ + + K +
Sbjct 643 HLDERDDSLDSKIGAILKKLTADKNAWPFASPVDVKEVPEYYDHIKHPIDFKTMQEKLKR 702
Query 64 RQYTSVAAFKDDINLIVSNCLLFNPVGSPSAWLRDRAVQLQQLAMQRLAEQ-PQIAEYEA 122
+ YT F D+N + NC +FN G+ + + + +L +LA++ L P+ + Y
Sbjct 703 KAYTHQHLFIADLNRLFQNCYVFN--GAEAVYYK-YGYKLNELALKLLKTSFPESSWYPE 759
Query 123 ALQQNAI 129
+Q +
Sbjct 760 LPEQKPL 766
> 7297294
Length=861
Score = 46.2 bits (108), Expect = 2e-05, Method: Composition-based stats.
Identities = 31/112 (27%), Positives = 54/112 (48%), Gaps = 5/112 (4%)
Query 5 MQQPIDALNVSFARVVNLLLQQQTFKPFVNRVDDRVAPGYSVVVQHPMFLRKVLSKCRER 64
++Q LN ++ L ++ + F V D +APGYS ++ PM + K +
Sbjct 273 LKQQKSPLNKLLEHLLRFLEKRDPHQFFAWPVTDDMAPGYSSIISRPMDFSTMRQKIDDH 332
Query 65 QYTSVAAFKDDINLIVSNCLLFNPVGSPSAWLRDRAVQLQQLAMQRLAEQPQ 116
+YT++ F DD L+ N + +N V + A +L Q+ M+ L QP+
Sbjct 333 EYTALTEFTDDFKLMCENAIKYNHVDTV---YNKAAKRLLQVGMKHL--QPE 379
> CE18461
Length=1430
Score = 45.1 bits (105), Expect = 4e-05, Method: Composition-based stats.
Identities = 21/65 (32%), Positives = 36/65 (55%), Gaps = 0/65 (0%)
Query 23 LLQQQTFKPFVNRVDDRVAPGYSVVVQHPMFLRKVLSKCRERQYTSVAAFKDDINLIVSN 82
L+ Q PF+ V+ ++ PGY +++ PM L+ + K + Y + F +DI L+ +N
Sbjct 1275 LVVQANALPFLEPVNPKLVPGYKMIISKPMDLKTIRQKNEKLIYETPEDFAEDIELMFAN 1334
Query 83 CLLFN 87
C FN
Sbjct 1335 CRQFN 1339
> CE18761
Length=1087
Score = 45.1 bits (105), Expect = 5e-05, Method: Composition-based stats.
Identities = 25/67 (37%), Positives = 36/67 (53%), Gaps = 3/67 (4%)
Query 44 YSVVVQHPMFLRKVLSKCRERQYTSVAAFKDDINLIVSNCLLFNPVGSPSAWLRDRAVQL 103
Y V+ +PM L+ + K +QY F DINL+V NC +NP GSP+ A++L
Sbjct 593 YLEVITNPMDLQTIKKKLDFKQYAEPEEFVHDINLMVDNCCKYNPKGSPA---HSNALEL 649
Query 104 QQLAMQR 110
+ QR
Sbjct 650 RSFFEQR 656
Score = 28.5 bits (62), Expect = 4.1, Method: Composition-based stats.
Identities = 15/46 (32%), Positives = 20/46 (43%), Gaps = 0/46 (0%)
Query 42 PGYSVVVQHPMFLRKVLSKCRERQYTSVAAFKDDINLIVSNCLLFN 87
P Y +V PM LR + + R Y D+N + NC FN
Sbjct 319 PEYHNIVNTPMDLRTIEKRLRNLYYWCAEDAIKDLNTLFDNCKKFN 364
> Hs4502453
Length=947
Score = 44.7 bits (104), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 22/75 (29%), Positives = 39/75 (52%), Gaps = 3/75 (4%)
Query 42 PGYSVVVQHPMFLRKVLSKCRERQYTSVAAFKDDINLIVSNCLLFNPVGSPSAWLRDRAV 101
P Y ++++PM L + + + Y + +D N + SNC L+N G + A
Sbjct 64 PDYYTIIKNPMDLNTIKKRLENKYYAKASECIEDFNTMFSNCYLYNKPGDDIVLM---AQ 120
Query 102 QLQQLAMQRLAEQPQ 116
L++L MQ+L++ PQ
Sbjct 121 ALEKLFMQKLSQMPQ 135
Score = 35.4 bits (80), Expect = 0.034, Method: Compositional matrix adjust.
Identities = 22/77 (28%), Positives = 37/77 (48%), Gaps = 5/77 (6%)
Query 17 ARVVNLLLQQQTFK---PFVNRVDDRVAP--GYSVVVQHPMFLRKVLSKCRERQYTSVAA 71
+ ++ +L ++ F PF N VD Y VV++PM L + K ++Y +
Sbjct 277 SEILKEMLAKKHFSYAWPFYNPVDVNALGLHNYYDVVKNPMDLGTIKEKMDNQEYKDAYS 336
Query 72 FKDDINLIVSNCLLFNP 88
F D+ L+ NC +NP
Sbjct 337 FAADVRLMFMNCYKYNP 353
> CE05993
Length=1250
Score = 44.3 bits (103), Expect = 8e-05, Method: Composition-based stats.
Identities = 25/67 (37%), Positives = 36/67 (53%), Gaps = 3/67 (4%)
Query 44 YSVVVQHPMFLRKVLSKCRERQYTSVAAFKDDINLIVSNCLLFNPVGSPSAWLRDRAVQL 103
Y V+ +PM L+ + K +QY F DINL+V NC +NP GSP+ A++L
Sbjct 633 YLEVITNPMDLQTIKKKLDFKQYAEPEEFVHDINLMVDNCCKYNPKGSPA---HSNALEL 689
Query 104 QQLAMQR 110
+ QR
Sbjct 690 RSFFEQR 696
Score = 33.9 bits (76), Expect = 0.12, Method: Composition-based stats.
Identities = 22/75 (29%), Positives = 33/75 (44%), Gaps = 3/75 (4%)
Query 42 PGYSVVVQHPMFLRKVLSKCRERQYTSVAAFKDDINLIVSNCLLFNPVGSPSAWLRDRAV 101
P Y +V PM LR + + R Y DIN + NC FNP P + A
Sbjct 319 PEYHNIVNTPMDLRTIEKRLRNLYYWCAEDAIKDINQVFINCYSFNP---PEYDVYKMAK 375
Query 102 QLQQLAMQRLAEQPQ 116
L++ + +L + P+
Sbjct 376 TLEKQVLSQLTQLPR 390
> YLR399c
Length=686
Score = 44.3 bits (103), Expect = 9e-05, Method: Composition-based stats.
Identities = 27/94 (28%), Positives = 48/94 (51%), Gaps = 5/94 (5%)
Query 31 PFVNRVD--DRVAPGYSVVVQHPMFLRKVLSKCRERQYTSVAAFKDDINLIVSNCLLFNP 88
PF+ VD P Y V+ PM L + K + QY ++ F+ D+ L+ NC FNP
Sbjct 339 PFLEPVDPVSMNLPTYFDYVKEPMDLGTIAKKLNDWQYQTMEDFERDVRLVFKNCYTFNP 398
Query 89 VGSPSAWLRDRAVQLQQLAMQRLAEQPQIAEYEA 122
G+ + R L+++ + A++P + +Y++
Sbjct 399 DGTIVNMMGHR---LEEVFNSKWADRPNLDDYDS 429
Score = 28.1 bits (61), Expect = 6.4, Method: Composition-based stats.
Identities = 23/88 (26%), Positives = 37/88 (42%), Gaps = 5/88 (5%)
Query 30 KPFVNRVD--DRVAPGYSVVVQHPMFLRKVLSKCRERQYTSVAAFKDDINLIVSNCLLFN 87
+PF+ VD P Y ++ PM L + K Y +D NL+V+N + FN
Sbjct 171 RPFLQPVDPVKLDIPFYFNYIKRPMDLSTIERKLNVGAYEVPEQITEDFNLMVNNSIKFN 230
Query 88 PVGSPSAWLRDRAVQLQQLAMQRLAEQP 115
P+A + A +Q + + P
Sbjct 231 ---GPNAGISQMARNIQASFEKHMLNMP 255
> Hs20357585_1
Length=1710
Score = 44.3 bits (103), Expect = 9e-05, Method: Composition-based stats.
Identities = 30/121 (24%), Positives = 60/121 (49%), Gaps = 13/121 (10%)
Query 3 AHMQQPIDAL-----NVSFARVVNLLLQQQTFK-----PFVNRVDDRVAPGYSVVVQHPM 52
A +++ I+ L V+F+ +++ ++ Q+ PF + V+ + P Y V+ +PM
Sbjct 1510 ARLEKAINPLLDDDDQVAFSFILDNIVTQKMMAVPDSWPFHHPVNKKFVPDYYKVIVNPM 1569
Query 53 FLRKVLSKCRERQYTSVAAFKDDINLIVSNCLLFNPVGSPSAWLRDRAVQLQQLAMQRLA 112
L + + +Y S +F DD+NLI++N + +N P + A ++ + Q L
Sbjct 1570 DLETIRKNISKHKYQSRESFLDDVNLILANSVKYN---GPESQYTKTAQEIVNVCYQTLT 1626
Query 113 E 113
E
Sbjct 1627 E 1627
Score = 39.3 bits (90), Expect = 0.003, Method: Composition-based stats.
Identities = 29/118 (24%), Positives = 53/118 (44%), Gaps = 4/118 (3%)
Query 8 PIDALNVSFARVVNLLLQQQTFKPFVNRVDDRVAPGYSVVVQHPMFLRKVLSKCRERQYT 67
P+ L+ ++N + PF V+ +V Y ++ PM L+ + R+R Y
Sbjct 1402 PMVTLSSILESIINDMRDLPNTYPFHTPVNAKVVKDYYKIITRPMDLQTLRENVRKRLYP 1461
Query 68 SVAAFKDDINLIVSNCLLFNPVGSPSAWLRDRAVQLQQLAMQRLAE-QPQIAEYEAAL 124
S F++ + LIV N +N P L + + L ++L E + ++A E A+
Sbjct 1462 SREEFREHLELIVKNSATYN---GPKHSLTQISQSMLDLCDEKLKEKEDKLARLEKAI 1516
> 7301065
Length=513
Score = 44.3 bits (103), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 20/51 (39%), Positives = 27/51 (52%), Gaps = 0/51 (0%)
Query 42 PGYSVVVQHPMFLRKVLSKCRERQYTSVAAFKDDINLIVSNCLLFNPVGSP 92
P Y VV+HPM L + + + Y + +D LI NCLL+N GSP
Sbjct 75 PDYHAVVKHPMDLSTIRKRLHNKYYWQASEALEDFKLIFDNCLLYNLEGSP 125
> Hs20357588_1
Length=1689
Score = 44.3 bits (103), Expect = 9e-05, Method: Composition-based stats.
Identities = 30/121 (24%), Positives = 60/121 (49%), Gaps = 13/121 (10%)
Query 3 AHMQQPIDAL-----NVSFARVVNLLLQQQTFK-----PFVNRVDDRVAPGYSVVVQHPM 52
A +++ I+ L V+F+ +++ ++ Q+ PF + V+ + P Y V+ +PM
Sbjct 1489 ARLEKAINPLLDDDDQVAFSFILDNIVTQKMMAVPDSWPFHHPVNKKFVPDYYKVIVNPM 1548
Query 53 FLRKVLSKCRERQYTSVAAFKDDINLIVSNCLLFNPVGSPSAWLRDRAVQLQQLAMQRLA 112
L + + +Y S +F DD+NLI++N + +N P + A ++ + Q L
Sbjct 1549 DLETIRKNISKHKYQSRESFLDDVNLILANSVKYN---GPESQYTKTAQEIVNVCYQTLT 1605
Query 113 E 113
E
Sbjct 1606 E 1606
Score = 39.3 bits (90), Expect = 0.003, Method: Composition-based stats.
Identities = 29/118 (24%), Positives = 53/118 (44%), Gaps = 4/118 (3%)
Query 8 PIDALNVSFARVVNLLLQQQTFKPFVNRVDDRVAPGYSVVVQHPMFLRKVLSKCRERQYT 67
P+ L+ ++N + PF V+ +V Y ++ PM L+ + R+R Y
Sbjct 1381 PMVTLSSILESIINDMRDLPNTYPFHTPVNAKVVKDYYKIITRPMDLQTLRENVRKRLYP 1440
Query 68 SVAAFKDDINLIVSNCLLFNPVGSPSAWLRDRAVQLQQLAMQRLAE-QPQIAEYEAAL 124
S F++ + LIV N +N P L + + L ++L E + ++A E A+
Sbjct 1441 SREEFREHLELIVKNSATYN---GPKHSLTQISQSMLDLCDEKLKEKEDKLARLEKAI 1495
> Hs7304919
Length=1674
Score = 43.9 bits (102), Expect = 9e-05, Method: Composition-based stats.
Identities = 26/98 (26%), Positives = 45/98 (45%), Gaps = 0/98 (0%)
Query 15 SFARVVNLLLQQQTFKPFVNRVDDRVAPGYSVVVQHPMFLRKVLSKCRERQYTSVAAFKD 74
+F ++V L++ PF+ V P Y +++ P+ L + K + +Y + F D
Sbjct 1555 AFEQLVVELVRHDDSWPFLKLVSKIQVPDYYDIIKKPIALNIIREKVNKCEYKLASEFID 1614
Query 75 DINLIVSNCLLFNPVGSPSAWLRDRAVQLQQLAMQRLA 112
DI L+ SNC +NP + A R + Q+L
Sbjct 1615 DIELMFSNCFEYNPRNTSEAKAGTRLQAFFHIQAQKLG 1652
> SPAC631.02
Length=727
Score = 43.9 bits (102), Expect = 1e-04, Method: Composition-based stats.
Identities = 30/89 (33%), Positives = 46/89 (51%), Gaps = 6/89 (6%)
Query 10 DALNVSFAR-VVNLLLQQQTFK---PFVNRVDDRVA--PGYSVVVQHPMFLRKVLSKCRE 63
DA + F + V+ LL++Q PF V+ P Y V++HPM L + +K
Sbjct 390 DAAEMKFCQSVLKELLKKQHEAYAYPFYKPVNPTACGCPDYFKVIKHPMDLGTMQNKLNH 449
Query 64 RQYTSVAAFKDDINLIVSNCLLFNPVGSP 92
+Y S+ AF+ D+ L+ NC FN G+P
Sbjct 450 NEYASMKAFEADMVLMFKNCYKFNSAGTP 478
Score = 41.2 bits (95), Expect = 7e-04, Method: Composition-based stats.
Identities = 23/64 (35%), Positives = 34/64 (53%), Gaps = 2/64 (3%)
Query 31 PFVNRVD--DRVAPGYSVVVQHPMFLRKVLSKCRERQYTSVAAFKDDINLIVSNCLLFNP 88
PF VD + P Y ++++P+ L + K Y+S F DD+NL+ SNC L+N
Sbjct 253 PFRAPVDPVKQNIPDYPTIIKNPIDLGTMQKKFSSGVYSSAQHFIDDMNLMFSNCFLYNG 312
Query 89 VGSP 92
SP
Sbjct 313 TESP 316
> CE25214
Length=851
Score = 43.9 bits (102), Expect = 1e-04, Method: Composition-based stats.
Identities = 21/70 (30%), Positives = 39/70 (55%), Gaps = 3/70 (4%)
Query 44 YSVVVQHPMFLRKVLSKCRERQYTSVAAFKDDINLIVSNCLLFNPVGSPSAWLRDRAVQL 103
Y +++ PM L+ + +K Y + F+ D+ L++ NC L+NPVG P + ++
Sbjct 299 YHKIIKEPMDLKSMKAKMESGAYKEPSDFEHDVRLMLRNCFLYNPVGDP---VHSFGLRF 355
Query 104 QQLAMQRLAE 113
Q++ +R AE
Sbjct 356 QEVFDRRWAE 365
Score = 30.4 bits (67), Expect = 1.1, Method: Composition-based stats.
Identities = 19/59 (32%), Positives = 25/59 (42%), Gaps = 2/59 (3%)
Query 31 PFVNRVDDRV--APGYSVVVQHPMFLRKVLSKCRERQYTSVAAFKDDINLIVSNCLLFN 87
PF VD P Y V PM L+ + ++ + YT DDI + NC FN
Sbjct 63 PFQKPVDAVALCIPLYHERVARPMDLKTIENRLKSTYYTCAQECIDDIETVFQNCYTFN 121
> 7298896
Length=2065
Score = 43.5 bits (101), Expect = 1e-04, Method: Composition-based stats.
Identities = 29/88 (32%), Positives = 43/88 (48%), Gaps = 2/88 (2%)
Query 8 PIDALNVSFARVVNLLLQQQTFKPFVNRVDDRVAPGYSVVVQHPMFLRKVLSKCRERQYT 67
P+ L+ + N L PF+ V + P Y VV PM L+ + R+R+YT
Sbjct 1471 PVVVLSSILEIIHNELRSMPDVSPFLFPVSAKKVPDYYRVVTKPMDLQTMREYIRQRRYT 1530
Query 68 SVAAFKDDINLIVSNCLLFNPVGSPSAW 95
S F +D+ IV N L++N G SA+
Sbjct 1531 SREMFLEDLKQIVDNSLIYN--GPQSAY 1556
Score = 39.3 bits (90), Expect = 0.003, Method: Composition-based stats.
Identities = 30/138 (21%), Positives = 58/138 (42%), Gaps = 5/138 (3%)
Query 11 ALNVSFARVVNLLLQQQTFKPFVNRVDDRVAPGYSVVVQHPMFLRKVLSKCRERQYTSVA 70
AL+ F ++ + + Q PF+ V+ + Y V++ PM L + +Y S A
Sbjct 1596 ALSFIFDKLHSQIKQLPESWPFLKPVNKKQVKDYYTVIKRPMDLETIGKNIEAHRYHSRA 1655
Query 71 AFKDDINLIVSNCLLFNPVGSPSAWLRDRAVQLQQLAMQRLAE-----QPQIAEYEAALQ 125
+ DI LI +NC +N + + ++ Q + +E + IA+ + +
Sbjct 1656 EYLADIELIATNCEQYNGSDTRYTKFSKKILEYAQTQLIEFSEHCGQLENNIAKTQERAR 1715
Query 126 QNAIHVQQQQQYSTYGYD 143
+NA + Y +D
Sbjct 1716 ENAPEFDEAWGNDDYNFD 1733
> Hs6382076
Length=832
Score = 43.5 bits (101), Expect = 1e-04, Method: Composition-based stats.
Identities = 23/82 (28%), Positives = 39/82 (47%), Gaps = 0/82 (0%)
Query 10 DALNVSFARVVNLLLQQQTFKPFVNRVDDRVAPGYSVVVQHPMFLRKVLSKCRERQYTSV 69
D L + ++ + Q+ PF+ V APGY V++ PM L+ + + + R Y S
Sbjct 726 DQLYSTLKSILQQVKSHQSAWPFMEPVKRTEAPGYYEVIRFPMDLKTMSERLKNRYYVSK 785
Query 70 AAFKDDINLIVSNCLLFNPVGS 91
F D+ + +NC +N S
Sbjct 786 KLFMADLQRVFTNCKEYNAAES 807
> YDL070w
Length=638
Score = 43.1 bits (100), Expect = 2e-04, Method: Composition-based stats.
Identities = 23/63 (36%), Positives = 34/63 (53%), Gaps = 2/63 (3%)
Query 31 PFVNRVDDRVA--PGYSVVVQHPMFLRKVLSKCRERQYTSVAAFKDDINLIVSNCLLFNP 88
PF+ VD P Y VV++PM L + + +Y ++ F DD+NL+ NC FNP
Sbjct 344 PFLQPVDPIALNLPNYFDVVKNPMDLGTISNNLMNWKYKTIDQFVDDLNLVFYNCFQFNP 403
Query 89 VGS 91
G+
Sbjct 404 EGN 406
Score = 38.9 bits (89), Expect = 0.003, Method: Composition-based stats.
Identities = 29/101 (28%), Positives = 47/101 (46%), Gaps = 5/101 (4%)
Query 30 KPFVNRVDDRV--APGYSVVVQHPMFLRKVLSKCRERQYTSVAAFKDDINLIVSNCLLFN 87
+PF+ VD P Y VQ PM L + +K + Y SV D +V NCL FN
Sbjct 156 RPFLKPVDPIALNIPHYFNYVQTPMDLSLIETKLQGNVYHSVEQVTSDFKTMVDNCLNFN 215
Query 88 PVGSPSAWLRDRAVQLQQLAMQRLAEQPQIAEYEAALQQNA 128
P + + A ++Q+ ++L+ P +AL++ +
Sbjct 216 ---GPESSISSMAKRIQKYFEKKLSAMPPRVLPASALKKTS 253
> ECU10g1430
Length=396
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 20/57 (35%), Positives = 31/57 (54%), Gaps = 0/57 (0%)
Query 31 PFVNRVDDRVAPGYSVVVQHPMFLRKVLSKCRERQYTSVAAFKDDINLIVSNCLLFN 87
PF+ VD P Y + PM L ++ K R +Y + AF D++L+V+NC +N
Sbjct 302 PFLRPVDPAEVPDYYKCIAKPMDLSTMVLKLRNNEYGCIEAFVADVHLMVNNCFEYN 358
> SPBC4B4.03
Length=803
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 21/62 (33%), Positives = 31/62 (50%), Gaps = 0/62 (0%)
Query 26 QQTFKPFVNRVDDRVAPGYSVVVQHPMFLRKVLSKCRERQYTSVAAFKDDINLIVSNCLL 85
+Q F PF D R+ P Y ++ PM L + K + +Y ++ F DD NL+ N
Sbjct 231 RQLFAPFERLPDPRMFPEYYQAIEQPMALEVIQKKLSKHRYETIEQFVDDFNLMFDNAKS 290
Query 86 FN 87
FN
Sbjct 291 FN 292
> Hs11067749
Length=726
Score = 42.7 bits (99), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 24/72 (33%), Positives = 35/72 (48%), Gaps = 3/72 (4%)
Query 44 YSVVVQHPMFLRKVLSKCRERQYTSVAAFKDDINLIVSNCLLFNPVGSPSAWLRDRAVQL 103
Y +++HPM L V K R+Y F D+ L+ SNC +NP P + A +L
Sbjct 348 YHDIIKHPMDLSTVKRKMDGREYPDAQGFAADVRLMFSNCYKYNP---PDHEVVAMARKL 404
Query 104 QQLAMQRLAEQP 115
Q + R A+ P
Sbjct 405 QDVFEMRFAKMP 416
Score = 38.1 bits (87), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 26/100 (26%), Positives = 48/100 (48%), Gaps = 5/100 (5%)
Query 19 VVNLLLQQQTFKPFVNRVD--DRVAPGYSVVVQHPMFLRKVLSKCRERQYTSVAAFKDDI 76
VV L + Q PF VD P Y ++++PM + + + Y S + D
Sbjct 46 VVKTLWKHQFAWPFYQPVDAIKLNLPDYHKIIKNPMDMGTIKKRLENNYYWSASECMQDF 105
Query 77 NLIVSNCLLFNPVGSPSAWLRDRAVQLQQLAMQRLAEQPQ 116
N + +NC ++N P+ + A L+++ +Q++A+ PQ
Sbjct 106 NTMFTNCYIYN---KPTDDIVLMAQALEKIFLQKVAQMPQ 142
> CE19384
Length=374
Score = 42.7 bits (99), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 20/62 (32%), Positives = 34/62 (54%), Gaps = 3/62 (4%)
Query 44 YSVVVQHPMFLRKVLSKCRERQYTSVAAFKDDINLIVSNCLLFNPVGSPSAWLRDRAVQL 103
Y V++ PM + + K +Y + FK+D L+++NCL +N G P A D A+Q
Sbjct 155 YHEVIKKPMDMSTIRKKLIGEEYDTAVEFKEDFKLMINNCLTYNNEGDPVA---DFALQF 211
Query 104 QQ 105
++
Sbjct 212 RK 213
> Hs17476222
Length=376
Score = 42.7 bits (99), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 17/52 (32%), Positives = 31/52 (59%), Gaps = 1/52 (1%)
Query 36 VDDRVAPGYSVVVQHPMFLRKVLSKCRERQYTSVAAFKDDINLIVSNCLLFN 87
V D +APGYS++++HP + K + Y S+ KD+ L+ +N +++N
Sbjct 233 VTDFIAPGYSMIIKHPRDFSTMKEKVKNNDYQSI-ELKDNFKLMCTNAMIYN 283
> Hs11426183
Length=837
Score = 42.4 bits (98), Expect = 3e-04, Method: Composition-based stats.
Identities = 23/79 (29%), Positives = 40/79 (50%), Gaps = 3/79 (3%)
Query 16 FARVVNLLLQQQTFK---PFVNRVDDRVAPGYSVVVQHPMFLRKVLSKCRERQYTSVAAF 72
+ + NLL Q ++ PF+ V AP Y V++ P+ L+ + + R R Y + F
Sbjct 734 YTTLKNLLAQIKSHPSAWPFMEPVKKSEAPDYYEVIRFPIDLKTMTERLRSRYYVTRKLF 793
Query 73 KDDINLIVSNCLLFNPVGS 91
D+ +++NC +NP S
Sbjct 794 VADLQRVIANCREYNPPDS 812
> Hs19718731
Length=1362
Score = 42.4 bits (98), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 24/76 (31%), Positives = 37/76 (48%), Gaps = 3/76 (3%)
Query 44 YSVVVQHPMFLRKVLSKCRERQYTSVAAFKDDINLIVSNCLLFNPVGSPSAWLRDRAVQL 103
Y +++HPM + + SK R+Y F D+ L+ SNC +NP P + A +L
Sbjct 390 YCDIIKHPMDMSTIKSKLEAREYRDAQEFGADVRLMFSNCYKYNP---PDHEVVAMARKL 446
Query 104 QQLAMQRLAEQPQIAE 119
Q + R A+ P E
Sbjct 447 QDVFEMRFAKMPDEPE 462
Score = 27.7 bits (60), Expect = 7.3, Method: Compositional matrix adjust.
Identities = 24/99 (24%), Positives = 43/99 (43%), Gaps = 5/99 (5%)
Query 19 VVNLLLQQQTFKPFVNRVD--DRVAPGYSVVVQHPMFLRKVLSKCRERQYTSVAAFKDDI 76
V+ L + Q PF VD P Y +++ PM + + + Y + D
Sbjct 70 VLKTLWKHQFAWPFQQPVDAVKLNLPDYYKIIKTPMDMGTIKKRLENNYYWNAQECIQDF 129
Query 77 NLIVSNCLLFNPVGSPSAWLRDRAVQLQQLAMQRLAEQP 115
N + +NC ++N G + A L++L +Q++ E P
Sbjct 130 NTMFTNCYIYNKPGDDIVLM---AEALEKLFLQKINELP 165
> Hs10835101
Length=837
Score = 42.4 bits (98), Expect = 3e-04, Method: Composition-based stats.
Identities = 23/79 (29%), Positives = 40/79 (50%), Gaps = 3/79 (3%)
Query 16 FARVVNLLLQQQTFK---PFVNRVDDRVAPGYSVVVQHPMFLRKVLSKCRERQYTSVAAF 72
+ + NLL Q ++ PF+ V AP Y V++ P+ L+ + + R R Y + F
Sbjct 734 YTTLKNLLAQIKSHPSAWPFMEPVKKSEAPDYYEVIRFPIDLKTMTERLRSRYYVTRKLF 793
Query 73 KDDINLIVSNCLLFNPVGS 91
D+ +++NC +NP S
Sbjct 794 VADLQRVIANCREYNPPDS 812
> Hs7657218
Length=722
Score = 42.4 bits (98), Expect = 3e-04, Method: Composition-based stats.
Identities = 24/76 (31%), Positives = 37/76 (48%), Gaps = 3/76 (3%)
Query 44 YSVVVQHPMFLRKVLSKCRERQYTSVAAFKDDINLIVSNCLLFNPVGSPSAWLRDRAVQL 103
Y +++HPM + + SK R+Y F D+ L+ SNC +NP P + A +L
Sbjct 390 YCDIIKHPMDMSTIKSKLEAREYRDAQEFGADVRLMFSNCYKYNP---PDHEVVAMARKL 446
Query 104 QQLAMQRLAEQPQIAE 119
Q + R A+ P E
Sbjct 447 QDVFEMRFAKMPDEPE 462
Score = 31.2 bits (69), Expect = 0.65, Method: Composition-based stats.
Identities = 24/99 (24%), Positives = 43/99 (43%), Gaps = 5/99 (5%)
Query 19 VVNLLLQQQTFKPFVNRVD--DRVAPGYSVVVQHPMFLRKVLSKCRERQYTSVAAFKDDI 76
V+ L + Q PF VD P Y +++ PM + + + Y + D
Sbjct 70 VLKTLWKHQFAWPFQQPVDAVKLNLPDYYKIIKTPMDMGTIKKRLENNYYWNAQECIQDF 129
Query 77 NLIVSNCLLFNPVGSPSAWLRDRAVQLQQLAMQRLAEQP 115
N + +NC ++N G + A L++L +Q++ E P
Sbjct 130 NTMFTNCYIYNKPGDDIVLM---AEALEKLFLQKINELP 165
> Hs14730464
Length=698
Score = 42.0 bits (97), Expect = 4e-04, Method: Composition-based stats.
Identities = 31/97 (31%), Positives = 44/97 (45%), Gaps = 6/97 (6%)
Query 19 VVNLLLQQQTFKPFVNRVDDRVAPGYSVVVQHPMFLRKVLSKCRERQYTSVAAFKDDINL 78
V L + F F V D Y V++ PM L V++K + Y + F DI+L
Sbjct 215 VTKRLATDKRFNIFSKPVSD-----YLEVIKEPMDLSTVITKIDKHNYLTAKDFLKDIDL 269
Query 79 IVSNCLLFNPVGSP-SAWLRDRAVQLQQLAMQRLAEQ 114
I SN L +NP P +R RA L+ A +A +
Sbjct 270 ICSNALEYNPDKDPGDKIIRHRACTLKDTAHAIIAAE 306
> Hs7304923
Length=1972
Score = 41.6 bits (96), Expect = 6e-04, Method: Composition-based stats.
Identities = 19/57 (33%), Positives = 30/57 (52%), Gaps = 0/57 (0%)
Query 31 PFVNRVDDRVAPGYSVVVQHPMFLRKVLSKCRERQYTSVAAFKDDINLIVSNCLLFN 87
PF+ V+ ++ PGY V++ PM + K QY ++ F D+ L+ NC FN
Sbjct 1888 PFLLPVNLKLVPGYKKVIKKPMDFSTIREKLSSGQYPNLETFALDVRLVFDNCETFN 1944
> SPAC1952.05
Length=454
Score = 41.2 bits (95), Expect = 7e-04, Method: Composition-based stats.
Identities = 21/57 (36%), Positives = 27/57 (47%), Gaps = 0/57 (0%)
Query 31 PFVNRVDDRVAPGYSVVVQHPMFLRKVLSKCRERQYTSVAAFKDDINLIVSNCLLFN 87
PF+ V P Y V++HPM L + + R QY SV F D I NC +N
Sbjct 367 PFMQPVSKEDVPDYYEVIEHPMDLSTMEFRLRNNQYESVEEFIRDAKYIFDNCRSYN 423
> HsM5730053
Length=920
Score = 41.2 bits (95), Expect = 7e-04, Method: Composition-based stats.
Identities = 24/82 (29%), Positives = 42/82 (51%), Gaps = 3/82 (3%)
Query 32 FVNRVDDRVAPGYSVVVQHPMFLRKVLSKCRERQYTSVAAFKDDINLIVSNCLLFNPVGS 91
F+ V D +APGY +VQ PM L + S A F+ DI L+ N +++N S
Sbjct 774 FLQPVTDDIAPGYHSIVQRPMDLSTIKKNIENGLIRSTAEFQRDIMLMFQNAVMYN---S 830
Query 92 PSAWLRDRAVQLQQLAMQRLAE 113
+ AV++Q+ ++++ +
Sbjct 831 SDHDVYHMAVEMQRDVLEQIQQ 852
> Hs21264560
Length=920
Score = 41.2 bits (95), Expect = 7e-04, Method: Composition-based stats.
Identities = 24/82 (29%), Positives = 42/82 (51%), Gaps = 3/82 (3%)
Query 32 FVNRVDDRVAPGYSVVVQHPMFLRKVLSKCRERQYTSVAAFKDDINLIVSNCLLFNPVGS 91
F+ V D +APGY +VQ PM L + S A F+ DI L+ N +++N S
Sbjct 774 FLQPVTDDIAPGYHSIVQRPMDLSTIKKNIENGLIRSTAEFQRDIMLMFQNAVMYN---S 830
Query 92 PSAWLRDRAVQLQQLAMQRLAE 113
+ AV++Q+ ++++ +
Sbjct 831 SDHDVYHMAVEMQRDVLEQIQQ 852
> Hs20555413
Length=1205
Score = 40.8 bits (94), Expect = 8e-04, Method: Composition-based stats.
Identities = 23/85 (27%), Positives = 39/85 (45%), Gaps = 0/85 (0%)
Query 3 AHMQQPIDALNVSFARVVNLLLQQQTFKPFVNRVDDRVAPGYSVVVQHPMFLRKVLSKCR 62
A M+ + NV ++LL ++ F V+ P Y + PM + K
Sbjct 585 AAMELELMPFNVLLRTTLDLLQEKDPAHIFAEPVNLSEVPDYLEFISKPMDFSTMRRKLE 644
Query 63 ERQYTSVAAFKDDINLIVSNCLLFN 87
Y ++ F++D NLIV+NC+ +N
Sbjct 645 SHLYRTLEEFEEDFNLIVTNCMKYN 669
> CE07224
Length=1042
Score = 40.8 bits (94), Expect = 9e-04, Method: Composition-based stats.
Identities = 16/45 (35%), Positives = 30/45 (66%), Gaps = 0/45 (0%)
Query 43 GYSVVVQHPMFLRKVLSKCRERQYTSVAAFKDDINLIVSNCLLFN 87
GY+ ++++P+ L+ + K +Y++VAA D+ L++SNC FN
Sbjct 542 GYTDIIENPICLKDMSEKAASGKYSTVAALSADVQLMLSNCATFN 586
> CE24921
Length=451
Score = 40.8 bits (94), Expect = 9e-04, Method: Compositional matrix adjust.
Identities = 19/61 (31%), Positives = 30/61 (49%), Gaps = 0/61 (0%)
Query 31 PFVNRVDDRVAPGYSVVVQHPMFLRKVLSKCRERQYTSVAAFKDDINLIVSNCLLFNPVG 90
PF N VD P Y ++ PM L + K +Y ++ F +D+N + N +NP G
Sbjct 311 PFRNPVDLNEFPDYEKFIKKPMDLSTITKKVERTEYLYLSQFVNDVNQMFENAKTYNPKG 370
Query 91 S 91
+
Sbjct 371 N 371
> CE15909
Length=405
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 19/61 (31%), Positives = 30/61 (49%), Gaps = 0/61 (0%)
Query 31 PFVNRVDDRVAPGYSVVVQHPMFLRKVLSKCRERQYTSVAAFKDDINLIVSNCLLFNPVG 90
PF N VD P Y ++ PM L + K +Y ++ F +D+N + N +NP G
Sbjct 265 PFRNPVDLNEFPDYEKFIKKPMDLSTITKKVERTEYLYLSQFVNDVNQMFENAKTYNPKG 324
Query 91 S 91
+
Sbjct 325 N 325
> CE15910
Length=510
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 19/61 (31%), Positives = 30/61 (49%), Gaps = 0/61 (0%)
Query 31 PFVNRVDDRVAPGYSVVVQHPMFLRKVLSKCRERQYTSVAAFKDDINLIVSNCLLFNPVG 90
PF N VD P Y ++ PM L + K +Y ++ F +D+N + N +NP G
Sbjct 370 PFRNPVDLNEFPDYEKFIKKPMDLSTITKKVERTEYLYLSQFVNDVNQMFENAKTYNPKG 429
Query 91 S 91
+
Sbjct 430 N 430
> At5g63330
Length=477
Score = 40.4 bits (93), Expect = 0.001, Method: Composition-based stats.
Identities = 24/75 (32%), Positives = 40/75 (53%), Gaps = 2/75 (2%)
Query 19 VVNLLLQQQTFKPFVNRVDDRV--APGYSVVVQHPMFLRKVLSKCRERQYTSVAAFKDDI 76
++N L ++ PF VD + P Y V++HPM L + S+ + +Y+S F D+
Sbjct 168 LLNRLWSHKSGWPFRTPVDPVMLNIPDYFNVIKHPMDLGTIRSRLCKGEYSSPLDFAADV 227
Query 77 NLIVSNCLLFNPVGS 91
L SN + +NP G+
Sbjct 228 RLTFSNSIAYNPPGN 242
> At1g05910
Length=1069
Score = 39.7 bits (91), Expect = 0.002, Method: Composition-based stats.
Identities = 25/86 (29%), Positives = 40/86 (46%), Gaps = 1/86 (1%)
Query 36 VDDRVAPGYSVVVQHPMFLRKVLSKCRERQYTSVAAFKDDINLIVSNCLLFNPVGSPSAW 95
V D AP Y ++Q PM +L + QY + F D++LIV N +N A
Sbjct 781 VTDEDAPNYRSIIQIPMDTATLLQRVDTGQYLTCTPFLQDVDLIVRNAKAYNGDDYAGAR 840
Query 96 LRDRAVQLQQLAMQRLAE-QPQIAEY 120
+ RA +L+ + L++ P + Y
Sbjct 841 IVSRAYELRDVVHGMLSQMDPALLTY 866
> 7294564
Length=813
Score = 39.7 bits (91), Expect = 0.002, Method: Composition-based stats.
Identities = 23/78 (29%), Positives = 38/78 (48%), Gaps = 0/78 (0%)
Query 10 DALNVSFARVVNLLLQQQTFKPFVNRVDDRVAPGYSVVVQHPMFLRKVLSKCRERQYTSV 69
+ L SFA V+ + Q T PF+ V P Y +++PM L+ + + ++ Y +
Sbjct 707 EKLATSFASVLQSVRQHTTAWPFLRPVTAAEVPDYYDHIKYPMDLKTMGERLKKGYYQTR 766
Query 70 AAFKDDINLIVSNCLLFN 87
F D+ I SNC +N
Sbjct 767 RLFMADMARIFSNCRFYN 784
> Hs4826806
Length=801
Score = 39.7 bits (91), Expect = 0.002, Method: Composition-based stats.
Identities = 24/72 (33%), Positives = 34/72 (47%), Gaps = 3/72 (4%)
Query 44 YSVVVQHPMFLRKVLSKCRERQYTSVAAFKDDINLIVSNCLLFNPVGSPSAWLRDRAVQL 103
Y +++HPM L V K R Y F D+ L+ SNC +NP P + A +L
Sbjct 386 YHDIIKHPMDLSTVKRKMENRDYRDAQEFAADVRLMFSNCYKYNP---PDHDVVAMARKL 442
Query 104 QQLAMQRLAEQP 115
Q + R A+ P
Sbjct 443 QDVFEFRYAKMP 454
Score = 35.0 bits (79), Expect = 0.050, Method: Composition-based stats.
Identities = 24/100 (24%), Positives = 46/100 (46%), Gaps = 5/100 (5%)
Query 19 VVNLLLQQQTFKPFVNRVD--DRVAPGYSVVVQHPMFLRKVLSKCRERQYTSVAAFKDDI 76
V+ L + Q PF VD P Y +++ PM + + + Y + + D
Sbjct 86 VMKALWKHQFAWPFRQPVDAVKLGLPDYHKIIKQPMDMGTIKRRLENNYYWAASECMQDF 145
Query 77 NLIVSNCLLFNPVGSPSAWLRDRAVQLQQLAMQRLAEQPQ 116
N + +NC ++N P+ + A L+++ +Q++A PQ
Sbjct 146 NTMFTNCYIYN---KPTDDIVLMAQTLEKIFLQKVASMPQ 182
> SPBC1734.15
Length=542
Score = 39.3 bits (90), Expect = 0.003, Method: Composition-based stats.
Identities = 19/53 (35%), Positives = 30/53 (56%), Gaps = 0/53 (0%)
Query 39 RVAPGYSVVVQHPMFLRKVLSKCRERQYTSVAAFKDDINLIVSNCLLFNPVGS 91
R P Y ++Q P+ + + +K + +Y S+ F DDI L+VSN +N GS
Sbjct 44 RYFPDYYQIIQKPICYKMMRNKAKTGKYLSMGDFYDDIRLMVSNAQTYNMPGS 96
Score = 36.2 bits (82), Expect = 0.024, Method: Composition-based stats.
Identities = 15/60 (25%), Positives = 34/60 (56%), Gaps = 0/60 (0%)
Query 32 FVNRVDDRVAPGYSVVVQHPMFLRKVLSKCRERQYTSVAAFKDDINLIVSNCLLFNPVGS 91
F++ R+ P Y +++ PM ++ + + ++ +YT++ +F D+N + N +N GS
Sbjct 170 FIDLPSKRLYPDYYEIIKSPMTIKMLEKRFKKGEYTTLESFVKDLNQMFINAKTYNAPGS 229
> CE05190
Length=636
Score = 38.9 bits (89), Expect = 0.003, Method: Composition-based stats.
Identities = 26/96 (27%), Positives = 45/96 (46%), Gaps = 3/96 (3%)
Query 19 VVNLLLQQQTFKPFVNRVDDRVAPGYSVVVQHPMFLRKVLSKCRERQYTSVAAFKDDINL 78
++ L+++ + F V +AP Y +++ PM L+ + + +Y S+ A K+D L
Sbjct 159 ILRKLVEKDPEQYFAFPVTPSMAPDYRDIIKTPMDLQTIRENIEDGKYASLPAMKEDCEL 218
Query 79 IVSNCLLFNPVGSPSAWLRDRAVQLQQLAMQRLAEQ 114
IVSN +N P+ A +L L EQ
Sbjct 219 IVSNAFQYN---QPNTVFYLAAKRLSNLIAYYFGEQ 251
> 7301208
Length=1654
Score = 38.9 bits (89), Expect = 0.003, Method: Composition-based stats.
Identities = 22/75 (29%), Positives = 40/75 (53%), Gaps = 3/75 (4%)
Query 39 RVAPGYSVVVQHPMFLRKVLSKCRERQYTSVAAFKDDINLIVSNCLLFNPVGSPSAWLRD 98
++ P Y V++HP+ LR + +K + Y+S+A + D+ + N LFN P + +
Sbjct 225 KIYPDYYDVIEHPIDLRLIATKIQMNAYSSLAEMERDLLQMTKNACLFN---EPGSQIYK 281
Query 99 RAVQLQQLAMQRLAE 113
A L+++ QR E
Sbjct 282 DAKSLKRIFTQRRIE 296
Lambda K H
0.322 0.132 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1712413322
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40