bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0987_orf1
Length=186
Score E
Sequences producing significant alignments: (Bits) Value
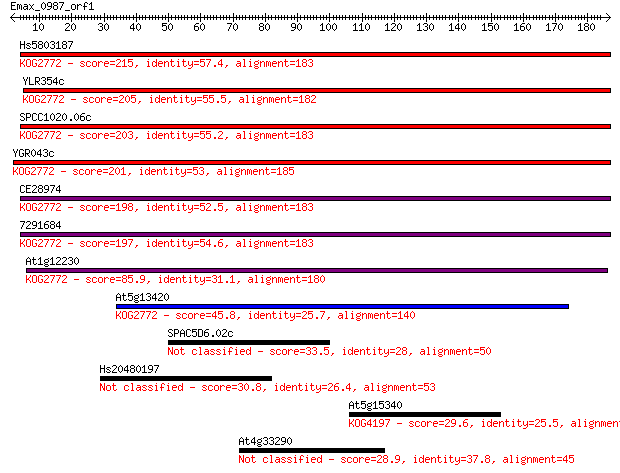
Hs5803187 215 4e-56
YLR354c 205 5e-53
SPCC1020.06c 203 2e-52
YGR043c 201 9e-52
CE28974 198 7e-51
7291684 197 7e-51
At1g12230 85.9 4e-17
At5g13420 45.8 5e-05
SPAC5D6.02c 33.5 0.26
Hs20480197 30.8 1.7
At5g15340 29.6 3.6
At4g33290 28.9 6.2
> Hs5803187
Length=337
Score = 215 bits (547), Expect = 4e-56, Method: Compositional matrix adjust.
Identities = 105/184 (57%), Positives = 143/184 (77%), Gaps = 3/184 (1%)
Query 4 TALDQLAKLSTIVADSADFEAIKEYNPVDATTNPTLVLQAVNNPKYKHLLDKAIKEAKEA 63
+ALDQL + +T+VAD+ DF AI EY P DATTNP+L+L A P Y+ L+++AI ++
Sbjct 13 SALDQLKQFTTVVADTGDFHAIDEYKPQDATTNPSLILAAAQMPAYQELVEEAIAYGRKL 72
Query 64 AKGKSQEIIIEEAIDRVLVLFGIEILQIIPGLVSTEIPADLSFNQQGSVERARRIIKLYE 123
G SQE I+ AID++ VLFG EIL+ IPG VSTE+ A LSF++ V RARR+I+LY+
Sbjct 73 --GGSQEDQIKNAIDKLFVLFGAEILKKIPGRVSTEVDARLSFDKDAMVARARRLIELYK 130
Query 124 EKGINKERILIKIAATWEGIQAAKELKKEN-INCNITLLFSLCQAIAASDAGAALVSPFV 182
E GI+K+RILIK+++TWEGIQA KEL++++ I+CN+TLLFS QA+A ++AG L+SPFV
Sbjct 131 EAGISKDRILIKLSSTWEGIQAGKELEEQHGIHCNMTLLFSFAQAVACAEAGVTLISPFV 190
Query 183 GRIL 186
GRIL
Sbjct 191 GRIL 194
> YLR354c
Length=335
Score = 205 bits (521), Expect = 5e-53, Method: Compositional matrix adjust.
Identities = 101/184 (54%), Positives = 142/184 (77%), Gaps = 4/184 (2%)
Query 5 ALDQL-AKLSTIVADSADFEAIKEYNPVDATTNPTLVLQAVNNPKYKHLLDKAIKEAKEA 63
+L+QL A + +VAD+ DF +I ++ P D+TTNP+L+L A P Y L+D A++ K+
Sbjct 15 SLEQLKASGTVVVADTGDFGSIAKFQPQDSTTNPSLILAAAKQPTYAKLIDVAVEYGKK- 73
Query 64 AKGKSQEIIIEEAIDRVLVLFGIEILQIIPGLVSTEIPADLSFNQQGSVERARRIIKLYE 123
GK+ E +E A+DR+LV FG EIL+I+PG VSTE+ A LSF+ Q ++E+AR IIKL+E
Sbjct 74 -HGKTTEEQVENAVDRLLVEFGKEILKIVPGRVSTEVDARLSFDTQATIEKARHIIKLFE 132
Query 124 EKGINKERILIKIAATWEGIQAAKEL-KKENINCNITLLFSLCQAIAASDAGAALVSPFV 182
++G++KER+LIKIA+TWEGIQAAKEL +K+ I+CN+TLLFS QA+A ++A L+SPFV
Sbjct 133 QEGVSKERVLIKIASTWEGIQAAKELEEKDGIHCNLTLLFSFVQAVACAEAQVTLISPFV 192
Query 183 GRIL 186
GRIL
Sbjct 193 GRIL 196
> SPCC1020.06c
Length=322
Score = 203 bits (516), Expect = 2e-52, Method: Compositional matrix adjust.
Identities = 101/184 (54%), Positives = 138/184 (75%), Gaps = 3/184 (1%)
Query 4 TALDQLAKLSTIV-ADSADFEAIKEYNPVDATTNPTLVLQAVNNPKYKHLLDKAIKEAKE 62
++L+QL T+V +D+ DFE+I +Y P DATTNP+L+L A P+Y L+D A+ AK
Sbjct 2 SSLEQLKATGTVVVSDTGDFESIAKYKPQDATTNPSLILAASKKPQYAALVDAAVDYAK- 60
Query 63 AAKGKSQEIIIEEAIDRVLVLFGIEILQIIPGLVSTEIPADLSFNQQGSVERARRIIKLY 122
AKG S IE A DR+L+ FG +IL I+PG VSTE+ A SF+ Q ++E+AR +IKLY
Sbjct 61 -AKGGSINSQIEIAFDRLLIEFGTKILAIVPGRVSTEVDARYSFDTQTTIEKARHLIKLY 119
Query 123 EEKGINKERILIKIAATWEGIQAAKELKKENINCNITLLFSLCQAIAASDAGAALVSPFV 182
E +GI +ER+LIKIA+T+EGIQAAK+L++E I+CN+TLLFS QA+A ++A L+SPFV
Sbjct 120 EAEGIGRERVLIKIASTYEGIQAAKQLEEEGIHCNLTLLFSFVQAVACAEANVTLISPFV 179
Query 183 GRIL 186
GRIL
Sbjct 180 GRIL 183
> YGR043c
Length=333
Score = 201 bits (510), Expect = 9e-52, Method: Compositional matrix adjust.
Identities = 98/187 (52%), Positives = 140/187 (74%), Gaps = 4/187 (2%)
Query 2 GTTALDQLAKLST-IVADSADFEAIKEYNPVDATTNPTLVLQAVNNPKYKHLLDKAIKEA 60
T++L+QL K T +VADS DFEAI +Y P D+TTNP+L+L A KY +D A++
Sbjct 12 ATSSLEQLKKAGTHVVADSGDFEAISKYEPQDSTTNPSLILAASKLEKYARFIDAAVEYG 71
Query 61 KEAAKGKSQEIIIEEAIDRVLVLFGIEILQIIPGLVSTEIPADLSFNQQGSVERARRIIK 120
++ GK+ IE A+D++LV FG +IL+++PG VSTE+ A LSF+++ +V++A IIK
Sbjct 72 RK--HGKTDHEKIENAMDKILVEFGTQILKVVPGRVSTEVDARLSFDKKATVKKALHIIK 129
Query 121 LYEEKGINKERILIKIAATWEGIQAAKELK-KENINCNITLLFSLCQAIAASDAGAALVS 179
LY++ G+ KER+LIKIA+TWEGIQAA+EL+ K I+CN+TLLFS QA+A ++A L+S
Sbjct 130 LYKDAGVPKERVLIKIASTWEGIQAARELEVKHGIHCNMTLLFSFTQAVACAEANVTLIS 189
Query 180 PFVGRIL 186
PFVGRI+
Sbjct 190 PFVGRIM 196
> CE28974
Length=319
Score = 198 bits (503), Expect = 7e-51, Method: Compositional matrix adjust.
Identities = 96/184 (52%), Positives = 138/184 (75%), Gaps = 3/184 (1%)
Query 4 TALDQLAKLSTIVADSADFEAIKEYNPVDATTNPTLVLQAVNNPKYKHLLDKAIKEAKEA 63
+ L+QL S +VAD+ DF AIKE+ P DATTNP+L+L A +Y L+D+++ AKE
Sbjct 2 SVLEQLKGASVVVADTGDFNAIKEFQPTDATTNPSLILAASKMEQYAALIDQSVAYAKEH 61
Query 64 AKGKSQEIIIEEAIDRVLVLFGIEILQIIPGLVSTEIPADLSFNQQGSVERARRIIKLYE 123
A G + +++ A+DR+ V+FG EIL+ IPG VSTE+ A LSF+ Q S++RA +I YE
Sbjct 62 ASGHQE--VLQAAMDRLFVVFGKEILKTIPGRVSTEVDARLSFDTQASIDRALGLIAQYE 119
Query 124 EKGINKERILIKIAATWEGIQAAKELK-KENINCNITLLFSLCQAIAASDAGAALVSPFV 182
++GI+K+RILIK+A+TWEGI+AAK L+ K I+CN+TLLF+ QA+A +++G L+SPFV
Sbjct 120 KEGISKDRILIKLASTWEGIRAAKFLESKHGIHCNMTLLFNFEQAVACAESGVTLISPFV 179
Query 183 GRIL 186
GRI+
Sbjct 180 GRIM 183
> 7291684
Length=320
Score = 197 bits (502), Expect = 7e-51, Method: Compositional matrix adjust.
Identities = 100/186 (53%), Positives = 140/186 (75%), Gaps = 7/186 (3%)
Query 4 TALDQLAKLSTIVADSADFEAIKEYNPVDATTNPTLVLQAVNNPKYKHLLDKAIKEAKEA 63
+ L +L K++TIVAD+ DFEAI Y P DATTNP+L+L A + +Y+ L+ KA+ E
Sbjct 2 SVLQELKKITTIVADTGDFEAINIYKPTDATTNPSLILSASSMERYQPLVQKAV----EY 57
Query 64 AKGKSQEII--IEEAIDRVLVLFGIEILQIIPGLVSTEIPADLSFNQQGSVERARRIIKL 121
AKGK + + EA+D + VLFG EIL+++PG VSTEI A LSF+ + SVE+A ++I L
Sbjct 58 AKGKKGSVSEQVAEAMDYLCVLFGTEILKVVPGRVSTEIDARLSFDTKKSVEKALKLIAL 117
Query 122 YEEKGINKERILIKIAATWEGIQAAKELKKEN-INCNITLLFSLCQAIAASDAGAALVSP 180
Y+ G++KERILIK+A+TWEGI+AA+ L+ E+ ++CN+TLLFS QA+A ++AG L+SP
Sbjct 118 YKSLGVDKERILIKLASTWEGIKAAEILENEHGVHCNLTLLFSFAQAVACAEAGVTLISP 177
Query 181 FVGRIL 186
FVGRIL
Sbjct 178 FVGRIL 183
> At1g12230
Length=377
Score = 85.9 bits (211), Expect = 4e-17, Method: Compositional matrix adjust.
Identities = 56/180 (31%), Positives = 97/180 (53%), Gaps = 27/180 (15%)
Query 6 LDQLAKLSTIVADSADFEAIKEYNPVDATTNPTLVLQAVNNPKYKHLLDKAIKEAKEAAK 65
L+ ++ S IV D+ F+ + + P AT + L+L P
Sbjct 71 LNAVSAFSEIVPDTVVFDDFERFPPTAATVSSALLLGICGLP------------------ 112
Query 66 GKSQEIIIEEAIDRVLVLFGIEILQIIPGLVSTEIPADLSFNQQGSVERARRIIKLYEEK 125
+ I R +V G ++++++PG VSTE+ A L+++ G + + +++LY E
Sbjct 113 ---------DTIFRAIVNVGGDLVKLVPGRVSTEVDARLAYDTNGIIRKVHDLLRLYNEI 163
Query 126 GINKERILIKIAATWEGIQAAKELKKENINCNITLLFSLCQAIAASDAGAALVSPFVGRI 185
+ +R+L KI ATW+GI+AA+ L+ E I ++T ++S QA AAS AGA+++ FVGR+
Sbjct 164 DVPHDRLLFKIPATWQGIEAARLLESEGIQTHMTFVYSFAQAAAASQAGASVIQIFVGRL 223
> At5g13420
Length=438
Score = 45.8 bits (107), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 36/141 (25%), Positives = 72/141 (51%), Gaps = 6/141 (4%)
Query 34 TTNPTLVLQAVNNPKYKHLLDKAIKEAKEAAKGKSQEIIIEEAIDRVLVLFGI-EILQII 92
T+NP + +A++ + + + E+ + + E+++++ D + I + +
Sbjct 112 TSNPAIFQKAISTSNAYNDQFRTLVESGKDIESAYWELVVKDIQDACKLFEPIYDQTEGA 171
Query 93 PGLVSTEIPADLSFNQQGSVERARRIIKLYEEKGINKERILIKIAATWEGIQAAKELKKE 152
G VS E+ L+ + QG+VE A+ Y K +N+ + IKI AT I + +++
Sbjct 172 DGYVSVEVSPRLADDTQGTVEAAK-----YLSKVVNRRNVYIKIPATAPCIPSIRDVIAA 226
Query 153 NINCNITLLFSLCQAIAASDA 173
I+ N+TL+FS+ + A DA
Sbjct 227 GISVNVTLIFSIARYEAVIDA 247
> SPAC5D6.02c
Length=300
Score = 33.5 bits (75), Expect = 0.26, Method: Compositional matrix adjust.
Identities = 14/50 (28%), Positives = 25/50 (50%), Gaps = 0/50 (0%)
Query 50 KHLLDKAIKEAKEAAKGKSQEIIIEEAIDRVLVLFGIEILQIIPGLVSTE 99
+HLLDK K + + + + I + + L F +ILQ +P S++
Sbjct 224 RHLLDKTAKRYHDLCEKRPYKYITTDLLSPSLTCFASDILQTVPEYTSSQ 273
> Hs20480197
Length=283
Score = 30.8 bits (68), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 14/53 (26%), Positives = 27/53 (50%), Gaps = 0/53 (0%)
Query 29 NPVDATTNPTLVLQAVNNPKYKHLLDKAIKEAKEAAKGKSQEIIIEEAIDRVL 81
+P+ P L ++ NNP + L +++ + + +K + Q IIE A V+
Sbjct 16 SPLTQNLPPLLCMEKDNNPSHPQSLGQSVLKNQSGSKAQGQLAIIEPAFMSVM 68
> At5g15340
Length=623
Score = 29.6 bits (65), Expect = 3.6, Method: Composition-based stats.
Identities = 12/47 (25%), Positives = 24/47 (51%), Gaps = 0/47 (0%)
Query 106 FNQQGSVERARRIIKLYEEKGINKERILIKIAATWEGIQAAKELKKE 152
+ + G V +RI + EEK + +++ WEG++ +E+ E
Sbjct 156 YGKCGLVSEVKRIFEELEEKSVVSWTVVLDTVVKWEGLERGREVFHE 202
> At4g33290
Length=430
Score = 28.9 bits (63), Expect = 6.2, Method: Compositional matrix adjust.
Identities = 17/45 (37%), Positives = 24/45 (53%), Gaps = 0/45 (0%)
Query 72 IIEEAIDRVLVLFGIEILQIIPGLVSTEIPADLSFNQQGSVERAR 116
IIEEA +R V G + +P LV + PA +Q S+E+ R
Sbjct 360 IIEEAGERAGVNCGSYVCSYVPSLVRIKKPAQGKRKRQSSLEKLR 404
Lambda K H
0.316 0.133 0.358
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3022542264
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40