bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0988_orf1
Length=101
Score E
Sequences producing significant alignments: (Bits) Value
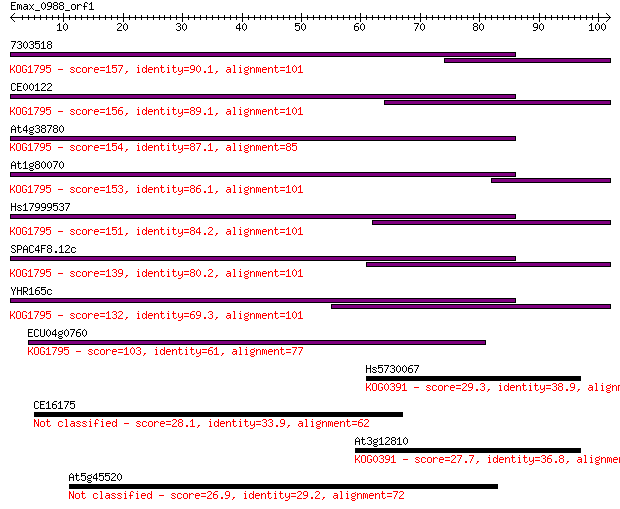
7303518 157 4e-39
CE00122 156 8e-39
At4g38780 154 5e-38
At1g80070 153 7e-38
Hs17999537 151 2e-37
SPAC4F8.12c 139 2e-33
YHR165c 132 2e-31
ECU04g0760 103 8e-23
Hs5730067 29.3 1.7
CE16175 28.1 4.1
At3g12810 27.7 4.8
At5g45520 26.9 8.3
> 7303518
Length=2396
Score = 157 bits (397), Expect = 4e-39, Method: Compositional matrix adjust.
Identities = 77/87 (88%), Positives = 79/87 (90%), Gaps = 2/87 (2%)
Query 1 FRVFKTTKFFQLTEIDWVEAGLQVSRQGYNMLNLLIHRKNLNYLHLDYNFNLRPVKTLTT 60
FR FK TKFFQ T +DWVEAGLQV RQGYNMLNLLIHRKNLNYLHLDYNFNL+PVKTLTT
Sbjct 533 FRSFKATKFFQTTTLDWVEAGLQVCRQGYNMLNLLIHRKNLNYLHLDYNFNLKPVKTLTT 592
Query 61 KERKKSRFGNAFHLCREILR--KLIKD 85
KERKKSRFGNAFHLCREILR KLI D
Sbjct 593 KERKKSRFGNAFHLCREILRLTKLIID 619
Score = 47.4 bits (111), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 19/28 (67%), Positives = 25/28 (89%), Gaps = 0/28 (0%)
Query 74 LCREILRKLIKDHGDMSSRKYRHDRRVY 101
+ E +RK+I+DHGDM+SRKYRHD+RVY
Sbjct 121 MPPEHIRKIIRDHGDMTSRKYRHDKRVY 148
> CE00122
Length=2329
Score = 156 bits (395), Expect = 8e-39, Method: Compositional matrix adjust.
Identities = 76/87 (87%), Positives = 79/87 (90%), Gaps = 2/87 (2%)
Query 1 FRVFKTTKFFQLTEIDWVEAGLQVSRQGYNMLNLLIHRKNLNYLHLDYNFNLRPVKTLTT 60
FR FK TKFFQ T +DWVEAGLQV RQGYNMLNLLIHRKNLNYLHLDYNFNL+PVKTLTT
Sbjct 465 FRSFKATKFFQTTTLDWVEAGLQVLRQGYNMLNLLIHRKNLNYLHLDYNFNLKPVKTLTT 524
Query 61 KERKKSRFGNAFHLCREILR--KLIKD 85
KERKKSRFGNAFHLCREILR KL+ D
Sbjct 525 KERKKSRFGNAFHLCREILRLTKLVVD 551
Score = 47.0 bits (110), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 22/42 (52%), Positives = 31/42 (73%), Gaps = 4/42 (9%)
Query 64 KKSRFGNA----FHLCREILRKLIKDHGDMSSRKYRHDRRVY 101
+K +FG + + E +RK+I+DHGDM+SRKYRHD+RVY
Sbjct 37 EKKKFGMSDTQKEEMPPEHVRKVIRDHGDMTSRKYRHDKRVY 78
> At4g38780
Length=2352
Score = 154 bits (388), Expect = 5e-38, Method: Compositional matrix adjust.
Identities = 74/87 (85%), Positives = 77/87 (88%), Gaps = 2/87 (2%)
Query 1 FRVFKTTKFFQLTEIDWVEAGLQVSRQGYNMLNLLIHRKNLNYLHLDYNFNLRPVKTLTT 60
FR TKFFQ TE+DWVE GLQV RQGYNMLNLLIHRKNLNYLHLDYNFNL+PVKTLTT
Sbjct 491 FRSLAATKFFQSTELDWVEVGLQVCRQGYNMLNLLIHRKNLNYLHLDYNFNLKPVKTLTT 550
Query 61 KERKKSRFGNAFHLCREILR--KLIKD 85
KERKKSRFGNAFHLCREILR KL+ D
Sbjct 551 KERKKSRFGNAFHLCREILRLTKLVVD 577
> At1g80070
Length=2382
Score = 153 bits (387), Expect = 7e-38, Method: Compositional matrix adjust.
Identities = 74/87 (85%), Positives = 77/87 (88%), Gaps = 2/87 (2%)
Query 1 FRVFKTTKFFQLTEIDWVEAGLQVSRQGYNMLNLLIHRKNLNYLHLDYNFNLRPVKTLTT 60
FR TKFFQ TE+DWVE GLQV RQGYNMLNLLIHRKNLNYLHLDYNFNL+PVKTLTT
Sbjct 519 FRSLAATKFFQSTELDWVEVGLQVCRQGYNMLNLLIHRKNLNYLHLDYNFNLKPVKTLTT 578
Query 61 KERKKSRFGNAFHLCREILR--KLIKD 85
KERKKSRFGNAFHLCREILR KL+ D
Sbjct 579 KERKKSRFGNAFHLCREILRLTKLVVD 605
Score = 39.3 bits (90), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 15/20 (75%), Positives = 19/20 (95%), Gaps = 0/20 (0%)
Query 82 LIKDHGDMSSRKYRHDRRVY 101
L +DHGDMSS+K+RHD+RVY
Sbjct 116 LARDHGDMSSKKFRHDKRVY 135
> Hs17999537
Length=2335
Score = 151 bits (382), Expect = 2e-37, Method: Compositional matrix adjust.
Identities = 73/87 (83%), Positives = 78/87 (89%), Gaps = 2/87 (2%)
Query 1 FRVFKTTKFFQLTEIDWVEAGLQVSRQGYNMLNLLIHRKNLNYLHLDYNFNLRPVKTLTT 60
FR FK TKFFQ T++DWVE LQV RQGYNMLNLLIHRKNLNYLHLDYNFNL+PVKTLTT
Sbjct 473 FRSFKATKFFQSTKLDWVEGWLQVCRQGYNMLNLLIHRKNLNYLHLDYNFNLKPVKTLTT 532
Query 61 KERKKSRFGNAFHLCREILR--KLIKD 85
KERKKSRFGNAFHLCRE+LR KL+ D
Sbjct 533 KERKKSRFGNAFHLCREVLRLTKLVVD 559
Score = 42.7 bits (99), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 20/42 (47%), Positives = 32/42 (76%), Gaps = 2/42 (4%)
Query 62 ERKKSRFGNA--FHLCREILRKLIKDHGDMSSRKYRHDRRVY 101
E++K F +A + E +R++I+DHGDM++RK+RHD+RVY
Sbjct 47 EKRKFGFVDAQKEDMPPEHVREIIRDHGDMTNRKFRHDKRVY 88
> SPAC4F8.12c
Length=2363
Score = 139 bits (349), Expect = 2e-33, Method: Compositional matrix adjust.
Identities = 70/87 (80%), Positives = 71/87 (81%), Gaps = 2/87 (2%)
Query 1 FRVFKTTKFFQLTEIDWVEAGLQVSRQGYNMLNLLIHRKNLNYLHLDYNFNLRPVKTLTT 60
R K TKFFQ T IDWVEAGLQV RQGYNML LLIHRK L YLHLDYN NL+P KTLTT
Sbjct 496 LRQLKNTKFFQSTSIDWVEAGLQVCRQGYNMLQLLIHRKGLTYLHLDYNCNLKPTKTLTT 555
Query 61 KERKKSRFGNAFHLCREILR--KLIKD 85
KERKKSRFGNAFHL REILR KLI D
Sbjct 556 KERKKSRFGNAFHLMREILRLTKLIVD 582
Score = 40.0 bits (92), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 22/51 (43%), Positives = 29/51 (56%), Gaps = 10/51 (19%)
Query 61 KERKKSRFG----------NAFHLCREILRKLIKDHGDMSSRKYRHDRRVY 101
+ +KS+FG L E LRK++KD GDMSSRK+R D+R Y
Sbjct 59 RASQKSKFGVKRKQGYVQTEKADLPPEHLRKIMKDRGDMSSRKFRADKRSY 109
> YHR165c
Length=2413
Score = 132 bits (332), Expect = 2e-31, Method: Compositional matrix adjust.
Identities = 64/87 (73%), Positives = 72/87 (82%), Gaps = 2/87 (2%)
Query 1 FRVFKTTKFFQLTEIDWVEAGLQVSRQGYNMLNLLIHRKNLNYLHLDYNFNLRPVKTLTT 60
+ K TK+FQ T IDWVEAGLQ+ RQG+NMLNLLIHRK L YLHLDYNFNL+P KTLTT
Sbjct 548 LKSLKNTKYFQQTTIDWVEAGLQLCRQGHNMLNLLIHRKGLTYLHLDYNFNLKPTKTLTT 607
Query 61 KERKKSRFGNAFHLCREILR--KLIKD 85
KERKKSR GN+FHL RE+L+ KLI D
Sbjct 608 KERKKSRLGNSFHLMRELLKMMKLIVD 634
Score = 33.5 bits (75), Expect = 0.098, Method: Compositional matrix adjust.
Identities = 14/47 (29%), Positives = 26/47 (55%), Gaps = 0/47 (0%)
Query 55 VKTLTTKERKKSRFGNAFHLCREILRKLIKDHGDMSSRKYRHDRRVY 101
K +T K ++ + + + E LRK+I H DM+S+ Y D++ +
Sbjct 115 AKKMTKKAKRSNLYTPKAEMPPEHLRKIINTHSDMASKMYNTDKKAF 161
> ECU04g0760
Length=2172
Score = 103 bits (257), Expect = 8e-23, Method: Compositional matrix adjust.
Identities = 47/77 (61%), Positives = 63/77 (81%), Gaps = 0/77 (0%)
Query 4 FKTTKFFQLTEIDWVEAGLQVSRQGYNMLNLLIHRKNLNYLHLDYNFNLRPVKTLTTKER 63
K T++FQ TEIDWVEAGLQ+ QG+ ML+ ++ RK L+YL LD+NFNL+P++ LTTKER
Sbjct 458 LKNTRYFQRTEIDWVEAGLQLVYQGHRMLSEVLRRKKLSYLVLDWNFNLKPIRQLTTKER 517
Query 64 KKSRFGNAFHLCREILR 80
KKSR G ++HL RE+L+
Sbjct 518 KKSRVGTSYHLTREMLK 534
> Hs5730067
Length=2971
Score = 29.3 bits (64), Expect = 1.7, Method: Composition-based stats.
Identities = 14/40 (35%), Positives = 22/40 (55%), Gaps = 4/40 (10%)
Query 61 KERKKSRFG----NAFHLCREILRKLIKDHGDMSSRKYRH 96
KERK R G NAFH+C + +++DH + +R+
Sbjct 506 KERKLKRQGWTKPNAFHVCITSYKLVLQDHQAFRRKNWRY 545
> CE16175
Length=327
Score = 28.1 bits (61), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 21/65 (32%), Positives = 30/65 (46%), Gaps = 4/65 (6%)
Query 5 KTTKFFQLTEIDWVEAGLQVSRQGYNMLNLLIHRKNLNYLHLDYNFNLRPVKT---LTTK 61
K + LT+I +Q+S G+N NL I LN L NF LR T +T
Sbjct 244 KVIPYILLTDIVITPLIIQISCLGFNKRNLGILLSTLNILQFS-NFVLRKTSTVHPMTIS 302
Query 62 ERKKS 66
E+ ++
Sbjct 303 EKSET 307
> At3g12810
Length=1048
Score = 27.7 bits (60), Expect = 4.8, Method: Composition-based stats.
Identities = 14/42 (33%), Positives = 23/42 (54%), Gaps = 4/42 (9%)
Query 59 TTKERKKSRFG----NAFHLCREILRKLIKDHGDMSSRKYRH 96
+ KERK R G N+FH+C R +I+D +K+++
Sbjct 204 SAKERKLKRQGWMKLNSFHVCITTYRLVIQDSKMFKRKKWKY 245
> At5g45520
Length=1167
Score = 26.9 bits (58), Expect = 8.3, Method: Composition-based stats.
Identities = 21/76 (27%), Positives = 36/76 (47%), Gaps = 5/76 (6%)
Query 11 QLTEIDWVEAGLQVSRQGYNMLNLLIHRKNLNYLHLDYNFNL----RPVKTLTTKERKKS 66
Q E D L++ + ++ ++ KNL D + NL R K + ++K S
Sbjct 85 QNIEEDQKNEDLEIRKLQNDIRQMIAAFKNLTQFQTDMSKNLERDLRSTKLIAILQKKNS 144
Query 67 RFGNAFHLCREILRKL 82
FG+ H+ +EI RK+
Sbjct 145 -FGSRSHVVKEIRRKV 159
Lambda K H
0.328 0.141 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1184494980
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40