bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0995_orf1
Length=122
Score E
Sequences producing significant alignments: (Bits) Value
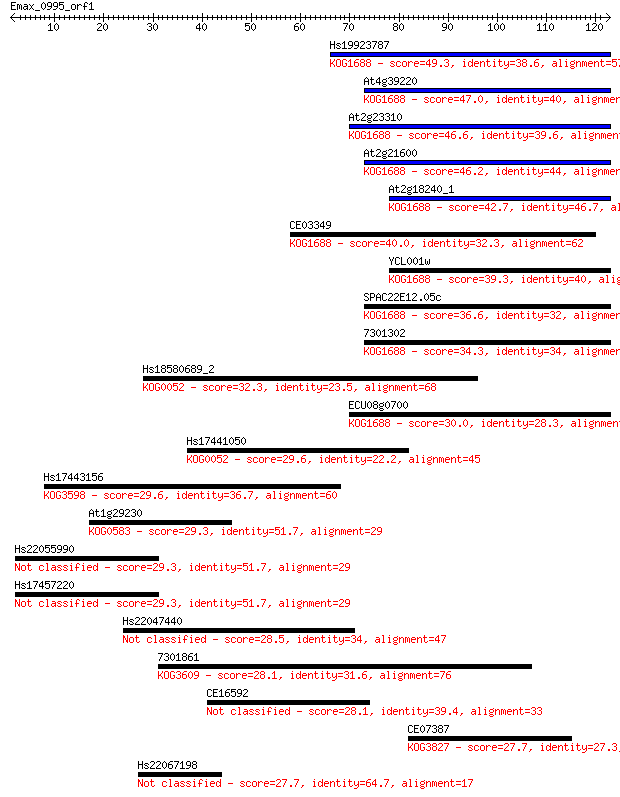
Hs19923787 49.3 2e-06
At4g39220 47.0 8e-06
At2g23310 46.6 1e-05
At2g21600 46.2 1e-05
At2g18240_1 42.7 2e-04
CE03349 40.0 0.001
YCL001w 39.3 0.002
SPAC22E12.05c 36.6 0.012
7301302 34.3 0.060
Hs18580689_2 32.3 0.22
ECU08g0700 30.0 1.0
Hs17441050 29.6 1.3
Hs17443156 29.6 1.5
At1g29230 29.3 2.0
Hs22055990 29.3 2.1
Hs17457220 29.3 2.1
Hs22047440 28.5 3.5
7301861 28.1 3.8
CE16592 28.1 3.9
CE07387 27.7 5.0
Hs22067198 27.7 5.2
> Hs19923787
Length=206
Score = 49.3 bits (116), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 22/57 (38%), Positives = 35/57 (61%), Gaps = 0/57 (0%)
Query 66 RVWASVGRVSNAYLNKSILYLKTRWLVLFACLAFYIARVYFLAGFFVVSYGLGIYLL 122
R + +G++ ++L+KS Y RW+V Y+ RVY L G+++V+Y LGIY L
Sbjct 20 RFFTRLGQIYQSWLDKSTPYTAVRWVVTLGLSFVYMIRVYLLQGWYIVTYALGIYHL 76
> At4g39220
Length=191
Score = 47.0 bits (110), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 20/50 (40%), Positives = 31/50 (62%), Gaps = 0/50 (0%)
Query 73 RVSNAYLNKSILYLKTRWLVLFACLAFYIARVYFLAGFFVVSYGLGIYLL 122
R+ YL+K+ + RW+ Y RVY++ GF++++YGLGIYLL
Sbjct 24 RIYQHYLDKTTPHANYRWIGTLVVALIYCLRVYYIQGFYIIAYGLGIYLL 73
> At2g23310
Length=211
Score = 46.6 bits (109), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 21/60 (35%), Positives = 38/60 (63%), Gaps = 7/60 (11%)
Query 70 SVGRVSNAY-------LNKSILYLKTRWLVLFACLAFYIARVYFLAGFFVVSYGLGIYLL 122
+V R+ +A+ L+K++ ++ RW+ + YI RVYF+ GF++++Y +GIYLL
Sbjct 36 AVNRLIHAFSQRQQHLLDKTVPHVLYRWIACLCVVLIYIVRVYFVEGFYIITYAIGIYLL 95
> At2g21600
Length=195
Score = 46.2 bits (108), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 22/50 (44%), Positives = 30/50 (60%), Gaps = 0/50 (0%)
Query 73 RVSNAYLNKSILYLKTRWLVLFACLAFYIARVYFLAGFFVVSYGLGIYLL 122
RV YL+K+ + RW+ Y RVY + GF+++SYGLGIYLL
Sbjct 24 RVYQYYLDKTTPHSTNRWIGTLVFFLIYCLRVYSIHGFYIISYGLGIYLL 73
> At2g18240_1
Length=180
Score = 42.7 bits (99), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 21/45 (46%), Positives = 28/45 (62%), Gaps = 0/45 (0%)
Query 78 YLNKSILYLKTRWLVLFACLAFYIARVYFLAGFFVVSYGLGIYLL 122
YL++S + RWLV YI RVY + G+FV+SYGL Y+L
Sbjct 33 YLDRSAPNIVRRWLVTLVAAVIYIYRVYSVYGYFVISYGLATYIL 77
> CE03349
Length=191
Score = 40.0 bits (92), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 20/63 (31%), Positives = 36/63 (57%), Gaps = 1/63 (1%)
Query 58 LADQP-FSSRVWASVGRVSNAYLNKSILYLKTRWLVLFACLAFYIARVYFLAGFFVVSYG 116
L D+P +SR + S+ YL++ + RW++ L F+ +R+ L GF++V+Y
Sbjct 5 LRDRPGVTSRFFHSLEVKYQYYLDRLTPHTAFRWVIALISLVFFASRIILLQGFYIVAYA 64
Query 117 LGI 119
+GI
Sbjct 65 VGI 67
> YCL001w
Length=188
Score = 39.3 bits (90), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 18/45 (40%), Positives = 28/45 (62%), Gaps = 0/45 (0%)
Query 78 YLNKSILYLKTRWLVLFACLAFYIARVYFLAGFFVVSYGLGIYLL 122
YL+K + K RW VL L ++ R+ G++V+ YGLG++LL
Sbjct 31 YLDKVTPHAKERWAVLGGLLCLFMVRITMAEGWYVICYGLGLFLL 75
> SPAC22E12.05c
Length=184
Score = 36.6 bits (83), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 16/50 (32%), Positives = 30/50 (60%), Gaps = 0/50 (0%)
Query 73 RVSNAYLNKSILYLKTRWLVLFACLAFYIARVYFLAGFFVVSYGLGIYLL 122
R+ +++++I Y RWL + +A + R+ + G+++V Y L IYLL
Sbjct 20 RLYRHWVDRTIPYTTYRWLTVSGLIALFFIRILLVRGWYIVCYTLAIYLL 69
> 7301302
Length=203
Score = 34.3 bits (77), Expect = 0.060, Method: Compositional matrix adjust.
Identities = 17/53 (32%), Positives = 30/53 (56%), Gaps = 3/53 (5%)
Query 73 RVSNAY---LNKSILYLKTRWLVLFACLAFYIARVYFLAGFFVVSYGLGIYLL 122
R+S Y L++S + + RW+ L ++ R++ G+++V Y LGIY L
Sbjct 20 RLSQTYQSALDRSTPHTRMRWVFAGFLLLLFVLRIFIYQGWYIVCYALGIYHL 72
> Hs18580689_2
Length=306
Score = 32.3 bits (72), Expect = 0.22, Method: Compositional matrix adjust.
Identities = 16/68 (23%), Positives = 34/68 (50%), Gaps = 2/68 (2%)
Query 28 RGGRNSEKGHLLGHYTIALRTMMASMSAEELADQPFSSRVWASVGRVSNAYLNKSILYLK 87
+ G+ E H L YT+ ++ ++ ++ + +QP+S + + + + N Y+ K L
Sbjct 50 KNGQTHE--HALLAYTLGVKQLIVGVNKMDSTEQPYSQKTYEEIVKEVNTYIMKIGCNLD 107
Query 88 TRWLVLFA 95
T VL +
Sbjct 108 TAAFVLIS 115
> ECU08g0700
Length=166
Score = 30.0 bits (66), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 15/53 (28%), Positives = 26/53 (49%), Gaps = 0/53 (0%)
Query 70 SVGRVSNAYLNKSILYLKTRWLVLFACLAFYIARVYFLAGFFVVSYGLGIYLL 122
+ + YL++ RW + FY R++ F++++Y LGIYLL
Sbjct 2 DLKTLQQIYLDRLAPRPDVRWGITGVLFLFYCIRIWSTGAFYLITYCLGIYLL 54
> Hs17441050
Length=462
Score = 29.6 bits (65), Expect = 1.3, Method: Composition-based stats.
Identities = 10/45 (22%), Positives = 26/45 (57%), Gaps = 0/45 (0%)
Query 37 HLLGHYTIALRTMMASMSAEELADQPFSSRVWASVGRVSNAYLNK 81
H L YT+ ++ ++ +++ ++ + P+SS + + + AY+ K
Sbjct 186 HTLLAYTLGMKQLIVTVNKMDITEPPYSSTCFEEISKEVKAYIKK 230
> Hs17443156
Length=1309
Score = 29.6 bits (65), Expect = 1.5, Method: Composition-based stats.
Identities = 22/66 (33%), Positives = 28/66 (42%), Gaps = 6/66 (9%)
Query 8 SEDRAGTLCGGLLPELPSTPRG------GRNSEKGHLLGHYTIALRTMMASMSAEELADQ 61
SE R L L+ LP++ +G G EKG LG + R S L+ Q
Sbjct 570 SERRGVWLVAPLIARLPTSVQGRVLKAAGEELEKGQHLGSSSKKERDRQKQKSMSLLSQQ 629
Query 62 PFSSRV 67
PF S V
Sbjct 630 PFLSLV 635
> At1g29230
Length=520
Score = 29.3 bits (64), Expect = 2.0, Method: Composition-based stats.
Identities = 15/35 (42%), Positives = 19/35 (54%), Gaps = 6/35 (17%)
Query 17 GGLLPELPSTPRGGRNS------EKGHLLGHYTIA 45
G + P+ P +PR RN+ E G LLGH T A
Sbjct 52 GNISPQSPRSPRSPRNNILMGKYELGKLLGHGTFA 86
> Hs22055990
Length=143
Score = 29.3 bits (64), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 15/29 (51%), Positives = 16/29 (55%), Gaps = 1/29 (3%)
Query 2 GFIRFLSEDRAGTLCGGLLPELPSTPRGG 30
GF L D+ LCG LLP LPS P G
Sbjct 20 GFPCLLQGDQDPPLCG-LLPHLPSDPHGA 47
> Hs17457220
Length=143
Score = 29.3 bits (64), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 15/29 (51%), Positives = 16/29 (55%), Gaps = 1/29 (3%)
Query 2 GFIRFLSEDRAGTLCGGLLPELPSTPRGG 30
GF L D+ LCG LLP LPS P G
Sbjct 20 GFPCLLQGDQDPPLCG-LLPHLPSDPHGA 47
> Hs22047440
Length=639
Score = 28.5 bits (62), Expect = 3.5, Method: Composition-based stats.
Identities = 16/47 (34%), Positives = 23/47 (48%), Gaps = 2/47 (4%)
Query 24 PSTPRGGRNSEKGHLLGHYTIALRTMMASMSAEELADQPFSSRVWAS 70
P++P GG +S HL I L + + A E+ DQP +W S
Sbjct 178 PASPGGGLDS--CHLNRSPAIPLHPNIGELQALEVLDQPEPGLIWES 222
> 7301861
Length=1275
Score = 28.1 bits (61), Expect = 3.8, Method: Composition-based stats.
Identities = 24/82 (29%), Positives = 37/82 (45%), Gaps = 6/82 (7%)
Query 31 RNSEKGHLLGHYTIALRTMMASMSAEEL----ADQPFS--SRVWASVGRVSNAYLNKSIL 84
R E+G L G +A+ T + ++ EEL +D F +W V +SN + IL
Sbjct 412 RKHERGSLPGPIELAIITYIMALIFEELKSLYSDGLFEYIMDLWNIVDYISNMFYVTWIL 471
Query 85 YLKTRWLVLFACLAFYIARVYF 106
T W+++ L F YF
Sbjct 472 CRATAWVIVHRDLWFRGIDPYF 493
> CE16592
Length=259
Score = 28.1 bits (61), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 13/33 (39%), Positives = 20/33 (60%), Gaps = 0/33 (0%)
Query 41 HYTIALRTMMASMSAEELADQPFSSRVWASVGR 73
HY A+ + S E A+QP+S +V++ VGR
Sbjct 107 HYLSAVNSSKIKKSFEISAEQPYSEQVFSGVGR 139
> CE07387
Length=514
Score = 27.7 bits (60), Expect = 5.0, Method: Composition-based stats.
Identities = 9/33 (27%), Positives = 22/33 (66%), Gaps = 0/33 (0%)
Query 82 SILYLKTRWLVLFACLAFYIARVYFLAGFFVVS 114
+++ +K RW +L+ L+F I+ +F +F+++
Sbjct 111 TVIEMKWRWCLLYFSLSFMISWSFFATVYFLIA 143
> Hs22067198
Length=402
Score = 27.7 bits (60), Expect = 5.2, Method: Composition-based stats.
Identities = 11/17 (64%), Positives = 12/17 (70%), Gaps = 0/17 (0%)
Query 27 PRGGRNSEKGHLLGHYT 43
P G +NS KG LLGH T
Sbjct 224 PFGSKNSSKGQLLGHET 240
Lambda K H
0.327 0.141 0.435
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1194805952
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40