bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
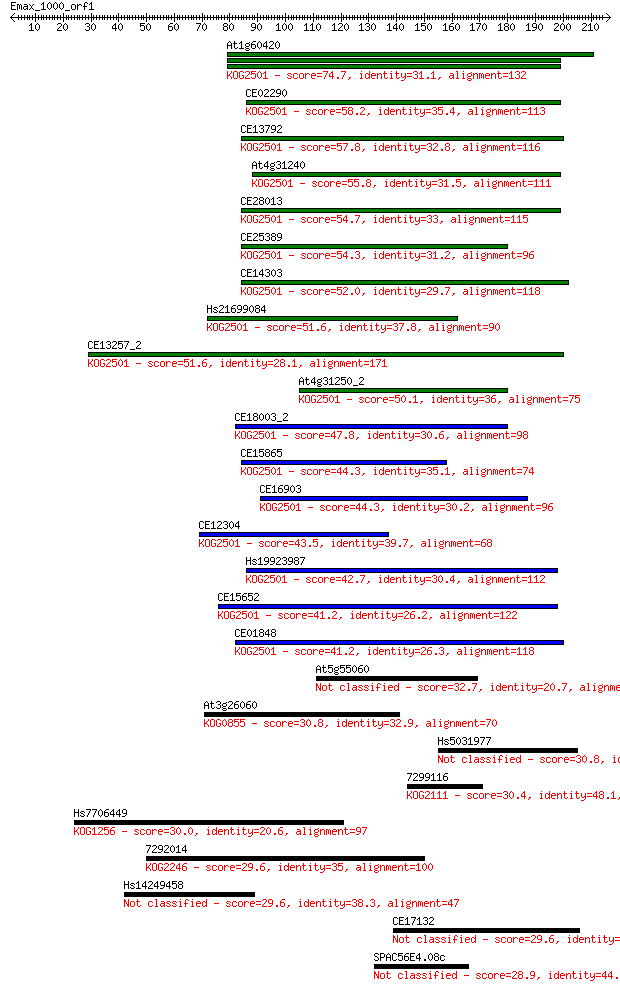
Query= Emax_1000_orf1
Length=216
Score E
Sequences producing significant alignments: (Bits) Value
At1g60420 74.7 1e-13
CE02290 58.2 1e-08
CE13792 57.8 2e-08
At4g31240 55.8 6e-08
CE28013 54.7 1e-07
CE25389 54.3 2e-07
CE14303 52.0 1e-06
Hs21699084 51.6 1e-06
CE13257_2 51.6 1e-06
At4g31250_2 50.1 3e-06
CE18003_2 47.8 2e-05
CE15865 44.3 2e-04
CE16903 44.3 2e-04
CE12304 43.5 4e-04
Hs19923987 42.7 5e-04
CE15652 41.2 0.002
CE01848 41.2 0.002
At5g55060 32.7 0.51
At3g26060 30.8 2.2
Hs5031977 30.8 2.4
7299116 30.4 2.9
Hs7706449 30.0 3.9
7292014 29.6 4.3
Hs14249458 29.6 4.8
CE17132 29.6 5.1
SPAC56E4.08c 28.9 8.6
> At1g60420
Length=578
Score = 74.7 bits (182), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 41/136 (30%), Positives = 71/136 (52%), Gaps = 20/136 (14%)
Query 79 NGNIVSQQHLIGKSVGLYFADSNSPKCRSFLPFLLQFYRTVNEGGSYQKIEVIFVSVDED 138
+G V L+GK++ +YF+ P CR+F P L++ Y+ + E + E+IF+S D D
Sbjct 352 DGAKVLVSDLVGKTILMYFSAHWCPPCRAFTPKLVEVYKQIKERN--EAFELIFISSDRD 409
Query 139 ESSYLDHRKHMPWLSIDFNNPLRTILLRHFRIGDEFTVPSLGEGHRAGLPALVVVGADG- 197
+ S+ ++ MPWL++ F +P + L + F++G G+P L +G G
Sbjct 410 QESFDEYYSQMPWLALPFGDPRKASLAKTFKVG--------------GIPMLAALGPTGQ 455
Query 198 ---RDAQFLQVGGGRD 210
++A+ L V G D
Sbjct 456 TVTKEARDLVVAHGAD 471
Score = 72.0 bits (175), Expect = 8e-13, Method: Compositional matrix adjust.
Identities = 38/120 (31%), Positives = 65/120 (54%), Gaps = 16/120 (13%)
Query 79 NGNIVSQQHLIGKSVGLYFADSNSPKCRSFLPFLLQFYRTVNEGGSYQKIEVIFVSVDED 138
+GN V L GK++GL F+ ++ KC P L++FY + E + + E++ +S+++D
Sbjct 192 DGNKVPVSELEGKTIGLLFSVASYRKCTELTPKLVEFYTKLKE--NKEDFEIVLISLEDD 249
Query 139 ESSYLDHRKHMPWLSIDFNNPLRTILLRHFRIGDEFTVPSLGEGHRAGLPALVVVGADGR 198
E S+ K PWL++ FN+ + L RHF + + LP LV++G DG+
Sbjct 250 EESFNQDFKTKPWLALPFNDKSGSKLARHFML--------------STLPTLVILGPDGK 295
Score = 66.6 bits (161), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 41/121 (33%), Positives = 63/121 (52%), Gaps = 18/121 (14%)
Query 79 NGNIVSQQHLIGKSVGLYFADSNSPKCRSFLPFLLQFYRTVNEGGSYQKIEVIFVSVDED 138
+G V L+GK +GLYF+ + C+ F P L++ Y NE S E++FVS DED
Sbjct 32 DGEQVKVDSLLGKKIGLYFSAAWCGPCQRFTPQLVEVY---NELSSKVGFEIVFVSGDED 88
Query 139 ESSYLDHRKHMPWLSIDF-NNPLRTILLRHFRIGDEFTVPSLGEGHRAGLPALVVVGADG 197
E S+ D+ + MPWL++ F ++ R L F++ G+P LV+V G
Sbjct 89 EESFGDYFRKMPWLAVPFTDSETRDRLDELFKV--------------RGIPNLVMVDDHG 134
Query 198 R 198
+
Sbjct 135 K 135
> CE02290
Length=149
Score = 58.2 bits (139), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 40/115 (34%), Positives = 59/115 (51%), Gaps = 11/115 (9%)
Query 86 QHLIGKSVGLYFADSNSPKCRSFLPFLLQFYRTVNEGGSYQKIEVIFVSVDEDESSYLDH 145
+HL GK VGLYF+ S P CR F P L +F+ + + + + EV+FVS D ++ ++
Sbjct 22 EHLKGKVVGLYFSASWCPPCRQFTPKLTRFFDEIRK--KHPEFEVVFVSRDREDGDLREY 79
Query 146 -RKHM-PWLSIDFNNPLRTILLRHFRIGDEFTVPSLGEGHRAGLPALVVVGADGR 198
+HM W +I F P LL + + T+PS+ R P VV D R
Sbjct 80 FLEHMGAWTAIPFGTPRIQELLEQYEVK---TIPSM----RIVKPNGDVVVQDAR 127
> CE13792
Length=155
Score = 57.8 bits (138), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 38/118 (32%), Positives = 53/118 (44%), Gaps = 21/118 (17%)
Query 84 SQQHLIGKSVGLYFADSNSPKCRSFLPFLLQFYRTVNEGGSYQKIEVIFVSVDEDESSYL 143
+ + L GK VG YF+ P CR F P L FY VNE + E++FVS D ES
Sbjct 33 ATEALAGKLVGFYFSAHWCPPCRGFTPILKDFYEEVNE-----EFEIVFVSSDRSESDLK 87
Query 144 DHRK--HMPWLSIDFNNPLRTILLRHFRIGDEFTVPSLGEGHRAGLPALVVVGADGRD 199
+ K H W I N + L + + +G+PAL++V DG +
Sbjct 88 MYMKECHGDWYHIPHGNGAKQKLSTKYGV--------------SGIPALIIVKPDGTE 131
> At4g31240
Length=204
Score = 55.8 bits (133), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 35/113 (30%), Positives = 55/113 (48%), Gaps = 19/113 (16%)
Query 88 LIGKSVGLYFADSNSPKCRSFLPFLLQFYRTV--NEGGSYQKIEVIFVSVDEDESSYLDH 145
L+GK++GLYF P RSF L+ Y + + GS+ EVI +S D D + +
Sbjct 13 LVGKTIGLYFGAHWCPPFRSFTSQLVDVYNELATTDKGSF---EVILISTDRDSREFNIN 69
Query 146 RKHMPWLSIDFNNPLRTILLRHFRIGDEFTVPSLGEGHRAGLPALVVVGADGR 198
+MPWL+I + + R L R F + +PALV++G + +
Sbjct 70 MTNMPWLAIPYEDRTRQDLCRIFNV--------------KLIPALVIIGPEEK 108
> CE28013
Length=179
Score = 54.7 bits (130), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 38/117 (32%), Positives = 61/117 (52%), Gaps = 11/117 (9%)
Query 84 SQQHLIGKSVGLYFADSNSPKCRSFLPFLLQFYRTVNEGGSYQKIEVIFVSVDEDESSYL 143
+++ L GK V LYF+ P C+ F P L++FY + + G + IEV+F S D ++
Sbjct 48 AEEALKGKVVALYFSAGWCPPCKQFTPKLVRFYHHLKKAG--KPIEVVFFSRDRSKADLE 105
Query 144 DH--RKHMPWLSIDFNNPLRTILLRHFRIGDEFTVPSLGEGHRAGLPALVVVGADGR 198
++ KH WL + + + + T F I T+P L + AG +VVV DG+
Sbjct 106 ENFTEKHGDWLCVKYGDDILTRYQSKFEIK---TIPVLRVINAAG--KMVVV--DGK 155
> CE25389
Length=149
Score = 54.3 bits (129), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 30/98 (30%), Positives = 55/98 (56%), Gaps = 7/98 (7%)
Query 84 SQQHLIGKSVGLYFADSNSPKCRSFLPFLLQFYRTVNEGGSYQKIEVIFVSVDEDESSYL 143
+++HL GK +GLYF+ S P CR+F P L +F+ + + ++ + E+IFVS D + S +
Sbjct 20 AEEHLKGKIIGLYFSASWCPPCRAFTPKLKEFFEEIKK--THPEFEIIFVSRDRNSSDLV 77
Query 144 DHRKHM--PWLSIDFNNPLRTILLRHFRIGDEFTVPSL 179
+ K W I F + L++ + + T+P++
Sbjct 78 TYFKEHQGEWTYIPFGSDKIMSLMQKYEVK---TIPAM 112
> CE14303
Length=122
Score = 52.0 bits (123), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 35/120 (29%), Positives = 50/120 (41%), Gaps = 21/120 (17%)
Query 84 SQQHLIGKSVGLYFADSNSPKCRSFLPFLLQFYRTVNEGGSYQKIEVIFVSVDEDESSYL 143
+ + L GK G YF+ P C F P L +FY V Y E++FVS D ES
Sbjct 9 ATEALAGKIGGFYFSAHWCPPCCMFTPILKKFYEKV-----YDDFEIVFVSSDPSESGLK 63
Query 144 DHRK--HMPWLSIDFNNPLRTILLRHFRIGDEFTVPSLGEGHRAGLPALVVVGADGRDAQ 201
+ + H W I F + + L + I G+P LV+V DG + +
Sbjct 64 KYMQECHGDWYYIPFGHEAKQKLCVKYEI--------------TGMPTLVIVKPDGTEVK 109
> Hs21699084
Length=135
Score = 51.6 bits (122), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 34/94 (36%), Positives = 43/94 (45%), Gaps = 4/94 (4%)
Query 72 EGSLRDCNGNIV-SQQHLIGKSVGLYFADSNSPKCRSFLPFLLQFYRT-VNEGGSYQKIE 129
E L C G V ++ L K V LYFA + R F P L FY V E E
Sbjct 7 ERHLVTCKGATVEAEAALQNKVVALYFAAARCAPSRDFTPLLCDFYTALVAEARRPAPFE 66
Query 130 VIFVSVDEDESSYLDHRK--HMPWLSIDFNNPLR 161
V+FVS D LD + H WL++ F++P R
Sbjct 67 VVFVSADGSCQEMLDFMRELHGAWLALPFHDPYR 100
> CE13257_2
Length=745
Score = 51.6 bits (122), Expect = 1e-06, Method: Composition-based stats.
Identities = 48/175 (27%), Positives = 75/175 (42%), Gaps = 28/175 (16%)
Query 29 KRRLNEEQAM--MATTAAAKQEAGAAAAAVTMPPAYRDMELILFPEGSLRDCNGNIVSQQ 86
KRR + +A+ + K E + P +R L P GS S++
Sbjct 576 KRREEKSRALKKLQKEETNKMEQMNNCTFLQNVPLFRH----LHPSGSY--------SER 623
Query 87 HLIGKSVGLYFADSNSPKCRSFLPFLLQFYRTVNEGGSYQKIEVIFVSVD--EDESSYLD 144
L GK +GLY++ R F P L QFY V+ + E++F+S D E E +Y
Sbjct 624 MLDGKVIGLYYSGYWCQPSRDFTPILAQFYSQVD-----KNFEILFISSDRSEQEMNYYL 678
Query 145 HRKHMPWLSIDFNNPLRTILLRHFRIGDEFTVPSLGEGHRAGLPALVVVGADGRD 199
H W + F++P+ + L+ F + +P+L P V+ DGRD
Sbjct 679 QSSHGDWFHLPFDSPI-SKHLQQFNTKN--AIPTL----IIIKPNGTVITVDGRD 726
> At4g31250_2
Length=281
Score = 50.1 bits (118), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 27/75 (36%), Positives = 42/75 (56%), Gaps = 5/75 (6%)
Query 105 CRSFLPFLLQFYRTVNEGGSYQKIEVIFVSVDEDESSYLDHRKHMPWLSIDFNNPLRTIL 164
C+ F P L++ Y + G +++E+IFVS D D +S+ +H MPWL++ FN L L
Sbjct 138 CKDFTPELIKLYENLQNRG--EELEIIFVSFDHDMTSFYEHFWCMPWLAVPFNLSLLNKL 195
Query 165 LRHFRIGDEFTVPSL 179
+ I +PSL
Sbjct 196 RDKYGIS---RIPSL 207
> CE18003_2
Length=144
Score = 47.8 bits (112), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 30/100 (30%), Positives = 47/100 (47%), Gaps = 10/100 (10%)
Query 82 IVSQQHLIGKSVGLYFADSNSPKCRSFLPFLLQFYRTVNEGGSYQKIEVIFVSVDEDESS 141
+ + + L GK VG+YF+ CR+F P L FY V + E++F S D+ ES
Sbjct 19 VDANEALAGKIVGIYFSAHWCGPCRNFTPVLKDFYEEVQDD-----FEIVFASSDQSESD 73
Query 142 YLDHRK--HMPWLSIDFNNPLRTILLRHFRIGDEFTVPSL 179
++ + H W I F N L + + T+P+L
Sbjct 74 LKNYMEECHGNWYYIPFGNDAEEKLSTKYDVS---TIPTL 110
> CE15865
Length=777
Score = 44.3 bits (103), Expect = 2e-04, Method: Composition-based stats.
Identities = 26/76 (34%), Positives = 37/76 (48%), Gaps = 7/76 (9%)
Query 84 SQQHLIGKSVGLYFADSNSPKCRSFLPFLLQFYRTVNEGGSYQKIEVIFVSVDED--ESS 141
+++ L GK VGLYF+ P R F P L QFY V + E++FVS D + E +
Sbjct 653 NERVLDGKVVGLYFSAHWCPPSRDFTPVLAQFYSQVED-----NFEILFVSSDNNTQEMN 707
Query 142 YLDHRKHMPWLSIDFN 157
+ H W + N
Sbjct 708 FYLQNFHGDWFHLPLN 723
> CE16903
Length=151
Score = 44.3 bits (103), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 29/97 (29%), Positives = 47/97 (48%), Gaps = 6/97 (6%)
Query 91 KSVGLYFADSNSPKCRSFLPFLLQFYRTVNEGGSYQKIEVIFVSVDEDESSYLDHRKH-M 149
K V LYF+ CR F P L +FY + G ++IEV+ VS D + L++ H
Sbjct 28 KVVALYFSAMWCGSCRQFTPKLKRFYEALKAAG--KEIEVVLVSRDREAEDLLEYLGHGG 85
Query 150 PWLSIDFNNPLRTILLRHFRIGDEFTVPSLGEGHRAG 186
W++I F + L+ + + T+P+ + AG
Sbjct 86 DWVAIPFGDERIQEYLKKYEVP---TIPAFKLINNAG 119
> CE12304
Length=174
Score = 43.5 bits (101), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 27/69 (39%), Positives = 37/69 (53%), Gaps = 3/69 (4%)
Query 69 LFPEGSLRDCNGNIVSQ-QHLIGKSVGLYFADSNSPKCRSFLPFLLQFYRTVNEGGSYQK 127
F SLR +G +V +HL GK V LYF+ S CR F P + + Y+ + + Q
Sbjct 5 FFSGTSLRLKDGTMVDAGEHLKGKIVVLYFSASWCGPCRQFTPIMKELYQQI--AATNQP 62
Query 128 IEVIFVSVD 136
IEVI +S D
Sbjct 63 IEVILLSRD 71
> Hs19923987
Length=212
Score = 42.7 bits (99), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 34/117 (29%), Positives = 54/117 (46%), Gaps = 19/117 (16%)
Query 86 QHLIGKSVGLYFADSNSPKCRSFLPFLLQFY-RTVNEGGSYQ--KIEVIFVSVDEDESSY 142
+ L + V L+F P+C++F+P L F+ R +E + ++ +++VS D E
Sbjct 28 RRLENRLVLLFFGAGACPQCQAFVPILKDFFVRLTDEFYVLRAAQLALVYVSQDSTEEQQ 87
Query 143 LDHRKHMP--WLSIDFNNPLRTILLRHFRIGDEFTVPSLGEGHRAGLPALVVVGADG 197
K MP WL + F + LR L R F + LPA+VV+ DG
Sbjct 88 DLFLKDMPKKWLFLPFEDDLRRDLGRQFSV--------------ERLPAVVVLKPDG 130
> CE15652
Length=178
Score = 41.2 bits (95), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 32/125 (25%), Positives = 62/125 (49%), Gaps = 19/125 (15%)
Query 76 RDCNGNIVSQQHLIGKSVGLYFADSNSPKCRSFLPFLLQFYRTVNEGGSYQKIEVIFVSV 135
R +GN+ + + K+V +YF+ C+ P + +FY V E + + +E+++VS
Sbjct 38 RGVDGNL-PKDYFENKTVVVYFSAGWCGSCKFLTPKIKKFYNAVKESDAGKNLEIVWVSK 96
Query 136 DEDESSYLD-HRKHMP-WLSIDFNNPLRTILLRHFRIGDEFTVPSLGEGHRA-GLPALVV 192
D++ + + + K++P W I F GDE + L E ++A +P L +
Sbjct 97 DKEAAHQEEYYEKNLPDWPYIPF--------------GDE-NIQKLSEKYKAVVIPVLKL 141
Query 193 VGADG 197
V ++G
Sbjct 142 VNSNG 146
> CE01848
Length=140
Score = 41.2 bits (95), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 31/118 (26%), Positives = 49/118 (41%), Gaps = 18/118 (15%)
Query 82 IVSQQHLIGKSVGLYFADSNSPKCRSFLPFLLQFYRTVNEGGSYQKIEVIFVSVDEDESS 141
+ + + L GK+VG YF+ P CR F P L FY V + + F + D
Sbjct 17 VDATEALAGKAVGFYFSAHWCPPCRGFTPILKDFYEEVEDEFEVVFVS--FDRSESDLKM 74
Query 142 YLDHRKHMPWLSIDFNNPLRTILLRHFRIGDEFTVPSLGEGHRAGLPALVVVGADGRD 199
Y+ +H W I + N L + + +G+PAL++V DG +
Sbjct 75 YM--SEHGDWYHIPYGNDAIKELSTKYGV--------------SGIPALIIVKPDGTE 116
> At5g55060
Length=645
Score = 32.7 bits (73), Expect = 0.51, Method: Compositional matrix adjust.
Identities = 12/58 (20%), Positives = 31/58 (53%), Gaps = 0/58 (0%)
Query 111 FLLQFYRTVNEGGSYQKIEVIFVSVDEDESSYLDHRKHMPWLSIDFNNPLRTILLRHF 168
F+++ + + +++ + + V E+ + + KH+PW+ +D N L++ LL +
Sbjct 212 FVVKLAEVIGSFTTPRRMALFWCRVVEELRRFWNEEKHIPWIPLDNNPDLKSCLLHQW 269
> At3g26060
Length=216
Score = 30.8 bits (68), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 23/71 (32%), Positives = 34/71 (47%), Gaps = 5/71 (7%)
Query 71 PEGSLRDCNGNIVSQQHLIGKSVGLYFADSN-SPKCRSFLPFLLQFYRTVNEGGSYQKIE 129
P+ +L+D NG VS + GK V LYF ++ +P C Y + G+ E
Sbjct 76 PDFTLKDQNGKPVSLKKYKGKPVVLYFYPADETPGCTKQACAFRDSYEKFKKAGA----E 131
Query 130 VIFVSVDEDES 140
VI +S D+ S
Sbjct 132 VIGISGDDSAS 142
> Hs5031977
Length=491
Score = 30.8 bits (68), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 18/50 (36%), Positives = 25/50 (50%), Gaps = 0/50 (0%)
Query 155 DFNNPLRTILLRHFRIGDEFTVPSLGEGHRAGLPALVVVGADGRDAQFLQ 204
D NPL T+L +G +F V +G++ P L V+ DG D LQ
Sbjct 313 DSGNPLDTVLKVLEILGKKFPVTENSKGYKLLPPYLRVIQGDGVDINTLQ 362
> 7299116
Length=340
Score = 30.4 bits (67), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 13/27 (48%), Positives = 17/27 (62%), Gaps = 0/27 (0%)
Query 144 DHRKHMPWLSIDFNNPLRTILLRHFRI 170
D K P +S+DFN P+R + LR RI
Sbjct 86 DDLKKSPAISLDFNQPVRAVRLRRDRI 112
> Hs7706449
Length=683
Score = 30.0 bits (66), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 20/103 (19%), Positives = 43/103 (41%), Gaps = 6/103 (5%)
Query 24 PYNAEKRRLNEEQAMMATTAAAKQEAGAAAAAVTMPPAYRDMELILFPEGSLRDCNGNIV 83
P++ + ++ E+ + + + G +PP+ D+ +I F G+ D G ++
Sbjct 214 PFDDDLKQRGEKSGIEILSLYDAENLGKEHFRKPVPPSPEDLSVICFTSGTTGDPKGAMI 273
Query 84 SQQHLIG------KSVGLYFADSNSPKCRSFLPFLLQFYRTVN 120
+ Q+++ K V + + S+LP F R V
Sbjct 274 THQNIVSNAAAFLKCVEHAYEPTPDDVAISYLPLAHMFERIVQ 316
> 7292014
Length=680
Score = 29.6 bits (65), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 35/121 (28%), Positives = 46/121 (38%), Gaps = 23/121 (19%)
Query 50 GAAAAAVTMPPAYRDMELILFPEGSLRDCNGNIVSQQHLIG---KSVGLYFADSNSPKCR 106
A V A R + L F N N S+ IG ++VG+ DS + R
Sbjct 182 SGGAGYVMSRDALRRLNLFAFNNSQFCPINNN--SEDRQIGFCLQNVGVVAGDSRDEEGR 239
Query 107 S-FLPFLLQF----------------YRTVNEGGSYQKIEVIFVSVDEDES-SYLDHRKH 148
FLP L+F Y VNE GS I +V + E E YL +R H
Sbjct 240 DRFLPLSLKFMLPTFPTDNWLPKLTFYEPVNETGSTSGISFHYVKIHEFEMYEYLLYRLH 299
Query 149 M 149
+
Sbjct 300 I 300
> Hs14249458
Length=1025
Score = 29.6 bits (65), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 18/47 (38%), Positives = 23/47 (48%), Gaps = 0/47 (0%)
Query 42 TAAAKQEAGAAAAAVTMPPAYRDMELILFPEGSLRDCNGNIVSQQHL 88
A A+ EAG A +PPA D L L+ E R G ++S Q L
Sbjct 2 VAMAEAEAGVAVEVRGLPPAVPDELLTLYFENRRRSGGGPVLSWQRL 48
> CE17132
Length=688
Score = 29.6 bits (65), Expect = 5.1, Method: Composition-based stats.
Identities = 19/67 (28%), Positives = 29/67 (43%), Gaps = 10/67 (14%)
Query 139 ESSYLDHRKHMPWLSIDFNNPLRTILLRHFRIGDEFTVPSLGEGHRAGLPALVVVGADGR 198
E++Y+ H ++ WL + F PL T+ L + + G GH L V R
Sbjct 17 EATYVHHDLNVVWLKLSFRVPLETLFLEYRQQG----------GHTLILSRQVYENILVR 66
Query 199 DAQFLQV 205
D F+ V
Sbjct 67 DTDFMSV 73
> SPAC56E4.08c
Length=378
Score = 28.9 bits (63), Expect = 8.6, Method: Compositional matrix adjust.
Identities = 15/34 (44%), Positives = 20/34 (58%), Gaps = 0/34 (0%)
Query 132 FVSVDEDESSYLDHRKHMPWLSIDFNNPLRTILL 165
+VSVDE ES + R P+LS F N ++LL
Sbjct 216 WVSVDEAESPHFTKRSPAPYLSKSFRNSALSLLL 249
Lambda K H
0.322 0.139 0.417
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4004762272
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40