bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_1022_orf1
Length=282
Score E
Sequences producing significant alignments: (Bits) Value
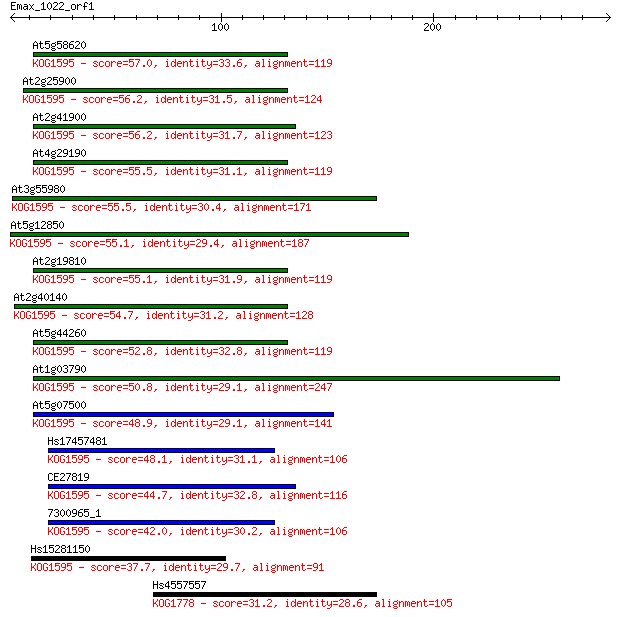
At5g58620 57.0 5e-08
At2g25900 56.2 7e-08
At2g41900 56.2 8e-08
At4g29190 55.5 1e-07
At3g55980 55.5 1e-07
At5g12850 55.1 2e-07
At2g19810 55.1 2e-07
At2g40140 54.7 2e-07
At5g44260 52.8 9e-07
At1g03790 50.8 3e-06
At5g07500 48.9 1e-05
Hs17457481 48.1 2e-05
CE27819 44.7 2e-04
7300965_1 42.0 0.002
Hs15281150 37.7 0.026
Hs4557557 31.2 2.8
> At5g58620
Length=607
Score = 57.0 bits (136), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 40/120 (33%), Positives = 54/120 (45%), Gaps = 14/120 (11%)
Query 12 YTLNEIELGTFRTQLCEQHRRSQCANPDACPHSH-CLTWQRRNPYEILYFPQLCPGIEFR 70
Y +E + F+ + C R+ + CP H +RR+P + Y CP EFR
Sbjct 205 YGTDEFRMYAFKIKPCS---RAYSHDWTECPFVHPGENARRRDPRKYHYSCVPCP--EFR 259
Query 71 RCNKKMNLVRHCNRGRFCSFAHSKEEELYHPLTYKTHLCSVFPNCLRHFCPFAHHTSELR 130
+ + C+RG C +AH E HP Y+T LC NC R C FAH ELR
Sbjct 260 KGS--------CSRGDTCEYAHGIFECWLHPAQYRTRLCKDETNCSRRVCFFAHKPEELR 311
> At2g25900
Length=315
Score = 56.2 bits (134), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 39/125 (31%), Positives = 57/125 (45%), Gaps = 14/125 (11%)
Query 7 PLSKHYTLNEIELGTFRTQLCEQHRRSQCANPDACPHSH-CLTWQRRNPYEILYFPQLCP 65
PL+ ++ +E + F+ + C + R CP +H +RR+P + Y CP
Sbjct 81 PLTDSFSSDEFRIYEFKIRRCARGRSHDWTE---CPFAHPGEKARRRDPRKFHYSGTACP 137
Query 66 GIEFRRCNKKMNLVRHCNRGRFCSFAHSKEEELYHPLTYKTHLCSVFPNCLRHFCPFAHH 125
EFR+ + C RG C F+H E HP Y+T C +C R C FAH
Sbjct 138 --EFRKGS--------CRRGDSCEFSHGVFECWLHPSRYRTQPCKDGTSCRRRICFFAHT 187
Query 126 TSELR 130
T +LR
Sbjct 188 TEQLR 192
> At2g41900
Length=727
Score = 56.2 bits (134), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 39/124 (31%), Positives = 54/124 (43%), Gaps = 14/124 (11%)
Query 12 YTLNEIELGTFRTQLCEQHRRSQCANPDACPHSH-CLTWQRRNPYEILYFPQLCPGIEFR 70
Y +E + +F+ + C R+ + CP H +RR+P + Y CP +FR
Sbjct 268 YATDEFRMYSFKVRPCS---RAYSHDWTECPFVHPGENARRRDPRKFHYSCVPCP--DFR 322
Query 71 RCNKKMNLVRHCNRGRFCSFAHSKEEELYHPLTYKTHLCSVFPNCLRHFCPFAHHTSELR 130
+ C RG C +AH E HP Y+T LC C R C FAH ELR
Sbjct 323 KG--------ACRRGDMCEYAHGVFECWLHPAQYRTRLCKDGTGCARRVCFFAHTPEELR 374
Query 131 DPFA 134
+A
Sbjct 375 PLYA 378
> At4g29190
Length=356
Score = 55.5 bits (132), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 37/120 (30%), Positives = 53/120 (44%), Gaps = 14/120 (11%)
Query 12 YTLNEIELGTFRTQLCEQHRRSQCANPDACPHSH-CLTWQRRNPYEILYFPQLCPGIEFR 70
Y+ + + F+ + C + R CP++H +RR+P + Y CP +FR
Sbjct 75 YSCDHFRMYDFKVRRCARGRSHDWTE---CPYAHPGEKARRRDPRKYHYSGTACP--DFR 129
Query 71 RCNKKMNLVRHCNRGRFCSFAHSKEEELYHPLTYKTHLCSVFPNCLRHFCPFAHHTSELR 130
+ C +G C FAH E HP Y+T C NCLR C FAH +LR
Sbjct 130 KGG--------CKKGDSCEFAHGVFECWLHPARYRTQPCKDGGNCLRKICFFAHSPDQLR 181
> At3g55980
Length=586
Score = 55.5 bits (132), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 52/181 (28%), Positives = 77/181 (42%), Gaps = 28/181 (15%)
Query 2 SASCPPLSKH-YTLNEIELGTFRTQLCEQ---HRRSQCANPDACPHSHCLTWQRRNPYEI 57
AS P +++ Y +E + +F+ + C + H ++CA ++ +RR+P +
Sbjct 202 DASLPDINEGVYGSDEFRMYSFKVKPCSRAYSHDWTECAFVHPGENA-----RRRDPRKY 256
Query 58 LYFPQLCPGIEFRRCNKKMNLVRHCNRGRFCSFAHSKEEELYHPLTYKTHLCSVFPNCLR 117
Y CP EFR+ + C +G C +AH E HP YKT LC C R
Sbjct 257 PYTCVPCP--EFRKGS--------CPKGDSCEYAHGVFESWLHPAQYKTRLCKDETGCAR 306
Query 118 HFCPFAHHTSELRDPFALPLIRALWGPQGPPI------GLQPTAATIAATPPTTTPAANA 171
C FAH E+R P A+ Q P GL P A + + P +P AN
Sbjct 307 KVCFFAHKREEMR-PVNASTGSAV--AQSPFSSLEMMPGLSPLAYSSGVSTPPVSPMANG 363
Query 172 A 172
Sbjct 364 V 364
> At5g12850
Length=706
Score = 55.1 bits (131), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 55/203 (27%), Positives = 84/203 (41%), Gaps = 30/203 (14%)
Query 1 ISASCPPL-SKHYTLNEIELGTFRTQLCEQHRRSQCANPDACPHSH-CLTWQRRNPYEIL 58
I S P + S Y+ +E + +F+ + C R+ + CP +H +RR+P +
Sbjct 241 IDPSLPDIKSGIYSTDEFRMFSFKIRPCS---RAYSHDWTECPFAHPGENARRRDPRKFH 297
Query 59 YFPQLCPGIEFRRCNKKMNLVRHCNRGRFCSFAHSKEEELYHPLTYKTHLCSVFPNCLRH 118
Y CP +F++ + C +G C +AH E HP Y+T LC C R
Sbjct 298 YTCVPCP--DFKKGS--------CKQGDMCEYAHGVFECWLHPAQYRTRLCKDGMGCNRR 347
Query 119 FCPFAHHTSELRDPF-----ALPLIRALWGPQGPPIGL---------QPTAATIAATPPT 164
C FAH ELR + LP RA + + P+AA + TPP
Sbjct 348 VCFFAHANEELRPLYPSTGSGLPSPRASSAVSASTMDMASVLNMLPGSPSAAQHSFTPP- 406
Query 165 TTPAANAAAAAGPQRMPLQTVEA 187
+P+ N + P Q + A
Sbjct 407 ISPSGNGSMPHSSMGWPQQNIPA 429
> At2g19810
Length=359
Score = 55.1 bits (131), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 38/121 (31%), Positives = 53/121 (43%), Gaps = 16/121 (13%)
Query 12 YTLNEIELGTFRTQLCEQHRRSQCANPDACPHSH-CLTWQRRNPYEILYFPQLCPGIEFR 70
YT + + F+ + C + R CP++H +RR+P + Y CP EFR
Sbjct 74 YTCDHFRMYEFKVRRCARGRSHDWTE---CPYAHPGEKARRRDPRKFHYSGTACP--EFR 128
Query 71 R-CNKKMNLVRHCNRGRFCSFAHSKEEELYHPLTYKTHLCSVFPNCLRHFCPFAHHTSEL 129
+ C C RG C F+H E HP Y+T C NC R C FAH ++
Sbjct 129 KGC---------CKRGDACEFSHGVFECWLHPARYRTQPCKDGGNCRRRVCFFAHSPDQI 179
Query 130 R 130
R
Sbjct 180 R 180
> At2g40140
Length=597
Score = 54.7 bits (130), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 40/130 (30%), Positives = 59/130 (45%), Gaps = 15/130 (11%)
Query 3 ASCPPLSKH-YTLNEIELGTFRTQLCEQHRRSQCANPDACPHSH-CLTWQRRNPYEILYF 60
AS P +++ Y ++ + +F+ + C R+ + CP H +RR+P + Y
Sbjct 200 ASLPDINEGVYGTDDFRMFSFKVKPCS---RAYSHDWTECPFVHPGENARRRDPRKYPYT 256
Query 61 PQLCPGIEFRRCNKKMNLVRHCNRGRFCSFAHSKEEELYHPLTYKTHLCSVFPNCLRHFC 120
CP EFR+ + C +G C +AH E HP Y+T LC C R C
Sbjct 257 CVPCP--EFRKGS--------CPKGDSCEYAHGVFESWLHPAQYRTRLCKDETGCARRVC 306
Query 121 PFAHHTSELR 130
FAH ELR
Sbjct 307 FFAHRRDELR 316
> At5g44260
Length=381
Score = 52.8 bits (125), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 39/120 (32%), Positives = 53/120 (44%), Gaps = 13/120 (10%)
Query 12 YTLNEIELGTFRTQLCEQHRRSQCANPDACPHSH-CLTWQRRNPYEILYFPQLCPGIEFR 70
Y + + F+ + C RS+ + CP SH +RR+P Y ++CP EF
Sbjct 56 YAGDHFRMYEFKIRRCT---RSRSHDWTDCPFSHPGEKARRRDPRRFHYTGEVCP--EFS 110
Query 71 RCNKKMNLVRHCNRGRFCSFAHSKEEELYHPLTYKTHLCSVFPNCLRHFCPFAHHTSELR 130
R C+RG C FAH E HP Y+T C +C R C FAH +LR
Sbjct 111 RHGD-------CSRGDECGFAHGVFECWLHPSRYRTEACKDGKHCKRKVCFFAHSPRQLR 163
> At1g03790
Length=393
Score = 50.8 bits (120), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 72/274 (26%), Positives = 103/274 (37%), Gaps = 42/274 (15%)
Query 12 YTLNEIELGTFRTQLCEQHRRSQCANPDACPHSH-CLTWQRRNPYEILYFPQLCPGIEFR 70
Y + + F+ + C RS+ + CP +H +RR+P Y ++CP EFR
Sbjct 78 YASDHFRMFEFKIRRCT---RSRSHDWTDCPFAHPGEKARRRDPRRFQYSGEVCP--EFR 132
Query 71 RCNKKMNLVRHCNRGRFCSFAHSKEEELYHPLTYKTHLCSVFPNCLRHFCPFAHHTSELR 130
R C+RG C FAH E HP+ Y+T C +C R C FAH +LR
Sbjct 133 RGGD-------CSRGDDCEFAHGVFECWLHPIRYRTEACKDGKHCKRKVCFFAHSPRQLR 185
Query 131 DPFALPLIRALWGPQGPPIGLQPTAATIAATPPTTT------------------PAANAA 172
LP P + ++ PT+T AN A
Sbjct 186 ---VLPPENVSGVSASPSPAAKNPCCLFCSSSPTSTLLGNLSHLSRSPSLSPPMSPANKA 242
Query 173 AAAGPQRMPLQTVEAASKGAAPLLLPDF-----GKIHKEAAAAAAAAASSSSTDSPQRQQ 227
AA R + +A+ A + D + + A A A+SSS +P
Sbjct 243 AAFSRLRNRAASAVSAAAAAGSMNYKDVLSELVNSLDSMSLAEALQASSSSPVTTPVSAA 302
Query 228 GQTQQ---ALQQQQTHQQQHQHDTALTTTVSPST 258
L Q+ H QQ Q + L +SPST
Sbjct 303 AAAFASSCGLSNQRLHLQQQQPSSPLQFALSPST 336
> At5g07500
Length=245
Score = 48.9 bits (115), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 41/149 (27%), Positives = 61/149 (40%), Gaps = 21/149 (14%)
Query 12 YTLNEIELGTFRTQLCEQHRRSQCANPDACPHSH-CLTWQRRNPYEILYFPQLCPGIEFR 70
Y +E + ++ + C R++ + CP++H RR+P Y CP FR
Sbjct 48 YGSDEFRMYAYKIKRCP---RTRSHDWTECPYAHRGEKATRRDPRRYTYCAVACPA--FR 102
Query 71 RCNKKMNLVRHCNRGRFCSFAHSKEEELYHPLTYKTHLCSVFPNCLRHFCPFAHHTSELR 130
C+RG C FAH E HP Y+T C+ C R C FAH +LR
Sbjct 103 NGA--------CHRGDSCEFAHGVFEYWLHPARYRTRACNAGNLCQRKVCFFAHAPEQLR 154
Query 131 DP-------FALPLIRALWGPQGPPIGLQ 152
+A +RA G G + ++
Sbjct 155 QSEGKHRCRYAYRPVRARGGGNGDGVTMR 183
> Hs17457481
Length=412
Score = 48.1 bits (113), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 33/110 (30%), Positives = 53/110 (48%), Gaps = 8/110 (7%)
Query 19 LGTFRTQLCEQHRRSQCANPDACPHSHCLTWQRRNPYEILYFPQLCPGIEFRRCNKKMNL 78
LG ++T+ C++ R C ACP+ H +RR+P + Y CP ++ +
Sbjct 213 LGNYKTEPCKKPPR-LCRQGYACPYYHNSKDRRRSPRKHKYRSSPCPNVKH---GDEWGD 268
Query 79 VRHCNRGRFCSFAHSKEEELYHPLTYKTHLCSVF---PNCLRH-FCPFAH 124
C G C + H++ E+ +HP YK+ C+ +C R FC FAH
Sbjct 269 PGKCENGDACQYCHTRTEQQFHPEIYKSTKCNDMQQSGSCPRGPFCAFAH 318
> CE27819
Length=704
Score = 44.7 bits (104), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 38/126 (30%), Positives = 58/126 (46%), Gaps = 17/126 (13%)
Query 19 LGTFRTQLCEQHRRSQCANPDACPHSHCLTWQRRNPYEILYFPQLCPGIEFRRCNKKMNL 78
L ++T+ C + R C ACP H +RR P Y CP + ++ ++
Sbjct 187 LSCYKTEQCRKPAR-LCRQGYACPFYHNSKDRRRPPALYKYRSTPCPAA--KTIDEWLD- 242
Query 79 VRHCNRGRFCSFAHSKEEELYHPLTYKTHLCSVFPNCLRH-------FCPFAHHTSEL-- 129
C G C + H++ E+ +HP YK+ C+ + L H FC FAHH SEL
Sbjct 243 PDICEAGDNCQYCHTRTEQQFHPEIYKSTKCN---DMLEHGYCPRAVFCAFAHHDSELHA 299
Query 130 -RDPFA 134
R+P+
Sbjct 300 HRNPYV 305
> 7300965_1
Length=541
Score = 42.0 bits (97), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 32/110 (29%), Positives = 52/110 (47%), Gaps = 8/110 (7%)
Query 19 LGTFRTQLCEQHRRSQCANPDACPHSHCLTWQRRNPYEILYFPQLCPGIEFRRCNKKMNL 78
L ++T+ C++ R C ACP H +RR+P + Y CP ++ ++
Sbjct 207 LANYKTEPCKRPPR-LCRQGYACPQYHNSKDKRRSPRKYKYRSTPCPNVKH---GEEWGE 262
Query 79 VRHCNRGRFCSFAHSKEEELYHPLTYKTHLCSVFPN---CLRH-FCPFAH 124
+C G C + H++ E+ +HP YK+ C+ C R FC FAH
Sbjct 263 PGNCEAGDNCQYCHTRTEQQFHPEIYKSTKCNDVQQAGYCPRSVFCAFAH 312
> Hs15281150
Length=756
Score = 37.7 bits (86), Expect = 0.026, Method: Compositional matrix adjust.
Identities = 27/91 (29%), Positives = 43/91 (47%), Gaps = 4/91 (4%)
Query 11 HYTLNEIELGTFRTQLCEQHRRSQCANPDACPHSHCLTWQRRNPYEILYFPQLCPGIEFR 70
H++ LG+++T+ C + R C ACPH H +RRNP Y CP ++
Sbjct 300 HFSDANFVLGSYKTEQCPKPPR-LCRQGYACPHYHNSRDRRRNPRRFQYRSTPCPSVKH- 357
Query 71 RCNKKMNLVRHCNRGRFCSFAHSKEEELYHP 101
+ C+ G C + HS+ E+ +HP
Sbjct 358 --GDEWGEPSRCDGGDGCQYCHSRTEQQFHP 386
> Hs4557557
Length=2414
Score = 31.2 bits (69), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 30/120 (25%), Positives = 45/120 (37%), Gaps = 24/120 (20%)
Query 68 EFRRCN--------KKMNLVRHCNRGRFCSFAHSKEEELYHPLTYKTHLCSVFPNCLRHF 119
E R+CN +N + HC G+ C AH + S + NC RH
Sbjct 360 EVRQCNLPHCRTMKNVLNHMTHCQSGKSCQVAHCASSR---------QIISHWKNCTRHD 410
Query 120 CPF---AHHTSELRD--PFALPLIRALWGPQGPPIGLQ--PTAATIAATPPTTTPAANAA 172
CP + + R+ P L P +G Q P +T++ P++ A AA
Sbjct 411 CPVCLPLKNAGDKRNQQPILTGAPVGLGNPSSLGVGQQSAPNLSTVSQIDPSSIERAYAA 470
Lambda K H
0.320 0.131 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 6198158050
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40