bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_1065_orf1
Length=146
Score E
Sequences producing significant alignments: (Bits) Value
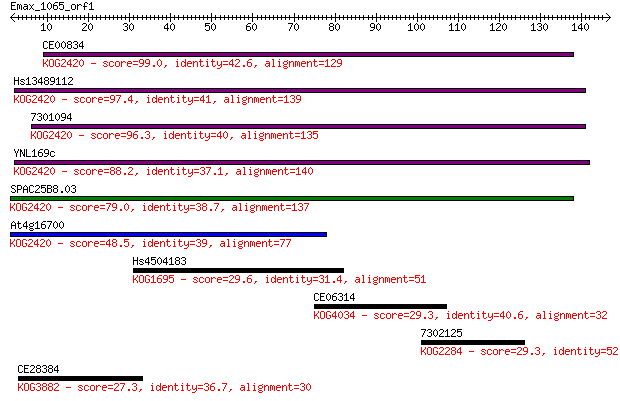
CE00834 99.0 3e-21
Hs13489112 97.4 8e-21
7301094 96.3 2e-20
YNL169c 88.2 6e-18
SPAC25B8.03 79.0 3e-15
At4g16700 48.5 5e-06
Hs4504183 29.6 2.1
CE06314 29.3 3.0
7302125 29.3 3.1
CE28384 27.3 9.7
> CE00834
Length=333
Score = 99.0 bits (245), Expect = 3e-21, Method: Compositional matrix adjust.
Identities = 55/129 (42%), Positives = 79/129 (61%), Gaps = 5/129 (3%)
Query 9 RHMHGECLPVFRSFLAKFNDIFSVQERIILSGSWIGGGLHIAAVAACNVGNIRLEKEPDL 68
RH+ G L V + L+ +F + ER++L+GSW G ++AVAA NVG+I ++ EP L
Sbjct 205 RHVPGLLLSVRPTLLSHVPHLFCLNERVVLNGSWRHGFFSMSAVAATNVGDIVVDAEPSL 264
Query 69 RTNQDRVVLRHLGGDVDIRTYLDKPLHFGVGSHVGEFRLGSTIVLIFEAPQEFEFSVAAG 128
RTN +V R ++ T + P + G VGEFRLGSTIVL+F+AP +F++ AG
Sbjct 265 RTN---IVRRKTQKIMNTETEIHAP--YVSGERVGEFRLGSTIVLVFQAPPTIKFAIKAG 319
Query 129 DKIRAGSRL 137
D +R G L
Sbjct 320 DPLRYGQSL 328
> Hs13489112
Length=375
Score = 97.4 bits (241), Expect = 8e-21, Method: Compositional matrix adjust.
Identities = 57/143 (39%), Positives = 78/143 (54%), Gaps = 8/143 (5%)
Query 2 DFTQTVRRHMHGECLPVFRSFLAKFNDIFSVQERIILSGSWIGGGLHIAAVAACNVGNIR 61
D+T + RRH G + V ++F ER++L+G W G + AV A NVG+IR
Sbjct 237 DWTVSHRRHFPGSLMSVNPGMARWIKELFCHNERVVLTGDWKHGFFSLTAVGATNVGSIR 296
Query 62 LEKEPDLRTNQDRVVLRHLGGDVDIRTYLDKPLHFGV----GSHVGEFRLGSTIVLIFEA 117
+ + DL TN R H G + +++ GV G H+GEF LGSTIVLIFEA
Sbjct 297 IYFDRDLHTNSPR----HSKGSYNDFSFVTHTNREGVPMRKGEHLGEFNLGSTIVLIFEA 352
Query 118 PQEFEFSVAAGDKIRAGSRLGGV 140
P++F F + G KIR G LG +
Sbjct 353 PKDFNFQLKTGQKIRFGEALGSL 375
> 7301094
Length=447
Score = 96.3 bits (238), Expect = 2e-20, Method: Composition-based stats.
Identities = 54/151 (35%), Positives = 75/151 (49%), Gaps = 24/151 (15%)
Query 6 TVRRHMHGECLPVFRSFLAKFNDIFSVQERIILSGSWIGGGLHIAAVAACNVGNIRLEKE 65
T+RRH GE L V +F + ER++ G W G AV A NVG++ + +
Sbjct 301 TIRRHFSGELLSVSPKVAGWLPGLFCLNERVLYMGQWKHGFFSYTAVGATNVGSVEIYMD 360
Query 66 PDLRTNQ----------------DRVVLRHLGGDVDIRTYLDKPLHFGVGSHVGEFRLGS 109
DL+TN+ D +VL + + P FG G VG+F +GS
Sbjct 361 ADLKTNRWTGFNVGKHPPSTYEYDELVLN--------KELTEAPKEFGKGDLVGQFNMGS 412
Query 110 TIVLIFEAPQEFEFSVAAGDKIRAGSRLGGV 140
TIVL+FEAP+ F+F + AG KIR G LG +
Sbjct 413 TIVLLFEAPKNFKFDIIAGQKIRVGESLGHI 443
> YNL169c
Length=500
Score = 88.2 bits (217), Expect = 6e-18, Method: Composition-based stats.
Identities = 52/146 (35%), Positives = 74/146 (50%), Gaps = 8/146 (5%)
Query 2 DFTQTVRRHMHGECLPVFRSFLAKFNDIFSVQERIILSGSWIGGGLHIAAVAACNVGNIR 61
D+ VRRH G+ V F F ++F + ER+ L GSW G + V A NVG+I+
Sbjct 352 DWVCKVRRHFPGDLFSVAPYFQRNFPNLFVLNERVALLGSWKYGFFSMTPVGATNVGSIK 411
Query 62 LEKEPDLRTNQDRVVLRHLGGDVDIRTYLDKP------LHFGVGSHVGEFRLGSTIVLIF 115
L + + TN +HL + + + G +G F LGST+VL F
Sbjct 412 LNFDQEFVTNSKSD--KHLEPHTCYQAVYENASKILGGMPLVKGEEMGGFELGSTVVLCF 469
Query 116 EAPQEFEFSVAAGDKIRAGSRLGGVG 141
EAP EF+F V GDK++ G +LG +G
Sbjct 470 EAPTEFKFDVRVGDKVKMGQKLGIIG 495
> SPAC25B8.03
Length=516
Score = 79.0 bits (193), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 53/143 (37%), Positives = 75/143 (52%), Gaps = 8/143 (5%)
Query 1 CDFTQTVRRHMHGECLPVFRSFLAKFNDIFSVQERIILSGSWIGGGLHIAAVAACNVGNI 60
D+ RRH GE V + +++F + ER+ L G + G + + V A NVG+I
Sbjct 376 ADWVIESRRHFSGELFSVSPFLARRLHNLFVLNERVALLGRYEHGFMSMIPVGATNVGSI 435
Query 61 RLEKEPDLRTNQDRVVLRH--LGGDVDIRTYLDKPLHFGV----GSHVGEFRLGSTIVLI 114
+ +P L TN R+VLR LG + P+ G+ G VG F+LGST+VL+
Sbjct 436 VINCDPTLSTN--RLVLRKKSLGTFQEAVYKNASPVLDGMPVSRGEQVGGFQLGSTVVLV 493
Query 115 FEAPQEFEFSVAAGDKIRAGSRL 137
FEAP +FEFS G +R G L
Sbjct 494 FEAPADFEFSTYQGQYVRVGEAL 516
> At4g16700
Length=434
Score = 48.5 bits (114), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 30/79 (37%), Positives = 43/79 (54%), Gaps = 2/79 (2%)
Query 1 CDFTQTVRRHMHGECL-PVFRSFLAKFN-DIFSVQERIILSGSWIGGGLHIAAVAACNVG 58
D+ TVRRH G P L N D + ++++L G W G + +AAV A N+G
Sbjct 318 ADWNATVRRHFAGLVYQPNMSGILLSPNIDHQLLIQQVVLEGIWKEGFMALAAVGATNIG 377
Query 59 NIRLEKEPDLRTNQDRVVL 77
+I L EP+LRTN+ + L
Sbjct 378 SIELFIEPELRTNKPKKKL 396
> Hs4504183
Length=210
Score = 29.6 bits (65), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 16/51 (31%), Positives = 25/51 (49%), Gaps = 4/51 (7%)
Query 31 SVQERIILSGSWIGGGLHIAAVAACNVGNIRLEKEPDLRTNQDRVVLRHLG 81
S +E ++ +W G L A+C G + ++ DL Q +LRHLG
Sbjct 28 SWKEEVVTVETWQEGSLK----ASCLYGQLPKFQDGDLTLYQSNTILRHLG 74
> CE06314
Length=223
Score = 29.3 bits (64), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 13/37 (35%), Positives = 23/37 (62%), Gaps = 5/37 (13%)
Query 75 VVLRHLGGDV-----DIRTYLDKPLHFGVGSHVGEFR 106
V +R++ GD+ D+R+YL++ L + SHV E +
Sbjct 165 VTIRNVDGDIFACENDLRSYLEEHLGHSIASHVDELK 201
> 7302125
Length=703
Score = 29.3 bits (64), Expect = 3.1, Method: Composition-based stats.
Identities = 13/25 (52%), Positives = 16/25 (64%), Gaps = 0/25 (0%)
Query 101 HVGEFRLGSTIVLIFEAPQEFEFSV 125
G + LGST V+ F PQEFE S+
Sbjct 473 QAGAWPLGSTQVIPFAVPQEFEKSI 497
> CE28384
Length=276
Score = 27.3 bits (59), Expect = 9.7, Method: Compositional matrix adjust.
Identities = 11/30 (36%), Positives = 17/30 (56%), Gaps = 0/30 (0%)
Query 3 FTQTVRRHMHGECLPVFRSFLAKFNDIFSV 32
+ T+ RH HG P++R F+ IFS+
Sbjct 217 YCNTIMRHSHGCITPLYRESQIHFSAIFSI 246
Lambda K H
0.325 0.144 0.442
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1748847648
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40