bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_1075_orf1
Length=118
Score E
Sequences producing significant alignments: (Bits) Value
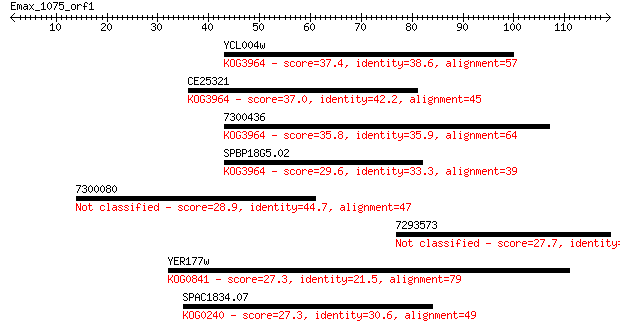
YCL004w 37.4 0.006
CE25321 37.0 0.008
7300436 35.8 0.021
SPBP18G5.02 29.6 1.6
7300080 28.9 2.3
7293573 27.7 5.4
YER177w 27.3 7.1
SPAC1834.07 27.3 7.7
> YCL004w
Length=521
Score = 37.4 bits (85), Expect = 0.006, Method: Composition-based stats.
Identities = 22/57 (38%), Positives = 35/57 (61%), Gaps = 1/57 (1%)
Query 43 MSLLGSSNLSVRSSVRDLELLCLLRTRDEQLVKQLQQEVYDVLLPFCVPAAAHVFSS 99
++++GSSN + R+ DLE L+ TRDE+L K+++ E+ D LL + P F S
Sbjct 445 ITVIGSSNYTRRAYSLDLESNALIITRDEELRKKMKAEL-DNLLQYTKPVTLEDFQS 500
> CE25321
Length=318
Score = 37.0 bits (84), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 19/50 (38%), Positives = 30/50 (60%), Gaps = 5/50 (10%)
Query 36 SRKIWAA-----MSLLGSSNLSVRSSVRDLELLCLLRTRDEQLVKQLQQE 80
++ +WA M+L+GSSN RS RDLE ++ TR+ L+ +L+ E
Sbjct 231 AKGLWAEHNNQLMTLIGSSNYGYRSVHRDLEAQVMVVTRNPTLIDRLKDE 280
> 7300436
Length=493
Score = 35.8 bits (81), Expect = 0.021, Method: Compositional matrix adjust.
Identities = 23/64 (35%), Positives = 32/64 (50%), Gaps = 1/64 (1%)
Query 43 MSLLGSSNLSVRSSVRDLELLCLLRTRDEQLVKQLQQEVYDVLLPFCVPAAAHVFSSRFP 102
++L+GSSN RS RDLE L T ++ L ++LQ E D L A + P
Sbjct 421 LTLIGSSNFGERSVNRDLETQVCLVTANKDLSQRLQAEA-DRLYDLSQTAEREIVQRPVP 479
Query 103 LWLQ 106
W+Q
Sbjct 480 RWVQ 483
> SPBP18G5.02
Length=534
Score = 29.6 bits (65), Expect = 1.6, Method: Composition-based stats.
Identities = 13/39 (33%), Positives = 25/39 (64%), Gaps = 0/39 (0%)
Query 43 MSLLGSSNLSVRSSVRDLELLCLLRTRDEQLVKQLQQEV 81
++ +GSSN + RS DLE ++ T++E+L ++ E+
Sbjct 463 LTTIGSSNYTSRSQQLDLESTLVVMTQNEKLKRKFSTEI 501
> 7300080
Length=472
Score = 28.9 bits (63), Expect = 2.3, Method: Composition-based stats.
Identities = 21/53 (39%), Positives = 27/53 (50%), Gaps = 9/53 (16%)
Query 14 LLHNSLTRNPASNSSCSSNC------VCSRKIWAAMSLLGSSNLSVRSSVRDL 60
L HNSL R P + SS S +C CS ++ SL+ SSN S + DL
Sbjct 367 LCHNSLPRQPPAKSSWSPDCEDRLTLECS---ISSPSLMRSSNDETCSELADL 416
> 7293573
Length=563
Score = 27.7 bits (60), Expect = 5.4, Method: Composition-based stats.
Identities = 12/42 (28%), Positives = 21/42 (50%), Gaps = 0/42 (0%)
Query 77 LQQEVYDVLLPFCVPAAAHVFSSRFPLWLQFAFNSLGLRSLL 118
+Q + +++P C H F +W+ FA +GL S+L
Sbjct 139 VQGFAFGLMVPTCYTTFNHYFVKNRVMWMSFAQTLIGLGSML 180
> YER177w
Length=267
Score = 27.3 bits (59), Expect = 7.1, Method: Compositional matrix adjust.
Identities = 17/80 (21%), Positives = 37/80 (46%), Gaps = 1/80 (1%)
Query 32 NCVCSRKI-WAAMSLLGSSNLSVRSSVRDLELLCLLRTRDEQLVKQLQQEVYDVLLPFCV 90
N + +R+ W +S + S S +EL+C R++ E + ++ ++ VL +
Sbjct 52 NVIGARRASWRIVSSIEQKEESKEKSEHQVELICSYRSKIETELTKISDDILSVLDSHLI 111
Query 91 PAAAHVFSSRFPLWLQFAFN 110
P+A S F ++ ++
Sbjct 112 PSATTGESKVFYYKMKGDYH 131
> SPAC1834.07
Length=554
Score = 27.3 bits (59), Expect = 7.7, Method: Composition-based stats.
Identities = 15/56 (26%), Positives = 28/56 (50%), Gaps = 7/56 (12%)
Query 35 CSRKIWAAMSLLGSSNLSVRSSVRDLELLCLLRT-------RDEQLVKQLQQEVYD 83
C +W+ L SNL+ +S++++ E++ RT RDE V + + +D
Sbjct 369 CHSNVWSGEHSLTLSNLAEKSNLKEAEIIQGNRTIQESNNDRDESTVASIHRHNFD 424
Lambda K H
0.324 0.132 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1167969826
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40