bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_1098_orf3
Length=355
Score E
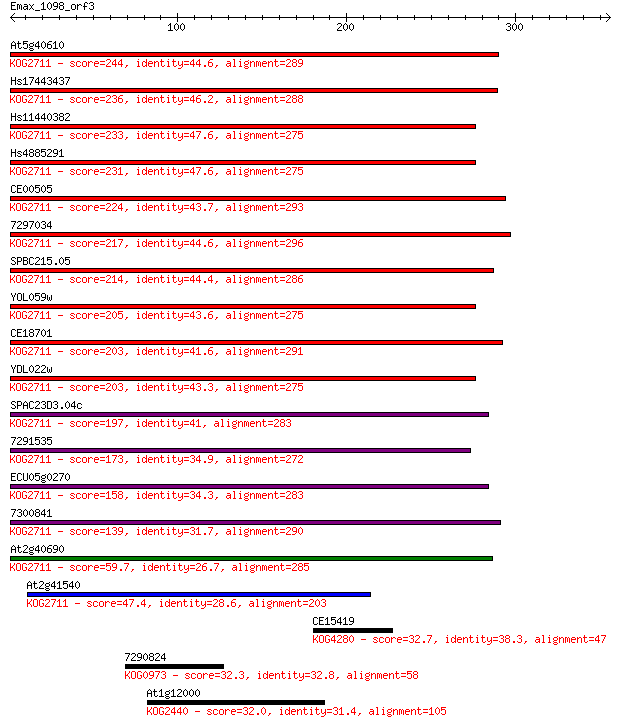
Sequences producing significant alignments: (Bits) Value
At5g40610 244 3e-64
Hs17443437 236 7e-62
Hs11440382 233 6e-61
Hs4885291 231 2e-60
CE00505 224 2e-58
7297034 217 4e-56
SPBC215.05 214 3e-55
YOL059w 205 1e-52
CE18701 203 4e-52
YDL022w 203 5e-52
SPAC23D3.04c 197 3e-50
7291535 173 5e-43
ECU05g0270 158 2e-38
7300841 139 1e-32
At2g40690 59.7 9e-09
At2g41540 47.4 5e-05
CE15419 32.7 1.3
7290824 32.3 1.5
At1g12000 32.0 2.1
> At5g40610
Length=400
Score = 244 bits (622), Expect = 3e-64, Method: Compositional matrix adjust.
Identities = 129/290 (44%), Positives = 184/290 (63%), Gaps = 6/290 (2%)
Query 1 YLPGFELPSNLRAVPSLTDACRDADLLVFVMPHQFIERACRELRKAKLLPPHCRAISLIK 60
YLPG +L N+ A P L +A +DA++LVFV PHQF++ C++L + AISL+K
Sbjct 115 YLPGIKLGRNVVADPDLENAVKDANMLVFVTPHQFMDGICKKLDGK--ITGDVEAISLVK 172
Query 61 GIGVRNGWPLTLTHEIGSILDLPKPCILSGANVANDVAAENFAEATIGHPDGETETAALW 120
G+ V+ P ++ I L + C+L GAN+AN++A E F+EAT+G+ G E A W
Sbjct 173 GMEVKKEGPCMISSLISKQLGI-NCCVLMGANIANEIAVEKFSEATVGY-RGSREIADTW 230
Query 121 QLLFDRPNFKINVLPDVDGVEVCGALKNVVALAGGFCEGMGQGTNTKAAILRLGVEEIKT 180
LF P F + + DV+GVE+CG LKNVVA+A GF +G+ G NTKAAI+R+G+ E+K
Sbjct 231 VQLFSTPYFMVTPVHDVEGVELCGTLKNVVAIAAGFVDGLEMGNNTKAAIMRIGLREMKA 290
Query 181 FTMLFFGDLLADTLYDSAGYADVITTVFGGRNARVAAAFVRQKGK-DWDQLEKEMLNGQK 239
+ L F + T ++S G ADVITT GGRN RVA AF + +GK +D+LE EML GQK
Sbjct 291 LSKLLFPSVKDSTFFESCGVADVITTCLGGRNRRVAEAFAKSRGKRSFDELEAEMLQGQK 350
Query 240 LQGPATLELVAEVIRANEVEHLFPLFTSTYAVAFQNADPQAILDVFRTNR 289
LQG +T V EV++ +FPLF++ + + P+AI+ +R N+
Sbjct 351 LQGVSTAREVYEVLKHCGWLEMFPLFSTVHQICTGRLQPEAIVQ-YRENK 399
> Hs17443437
Length=351
Score = 236 bits (601), Expect = 7e-62, Method: Compositional matrix adjust.
Identities = 133/293 (45%), Positives = 185/293 (63%), Gaps = 15/293 (5%)
Query 1 YLPGFELPSNLRAVPSLTDACRDADLLVFVMPHQFIERACRELRKAKLLPPHCRAISLIK 60
YLPG +LP N+ A+ +L++A +DADLLVFV+PHQFI R C E+ +P I+LIK
Sbjct 65 YLPGHKLPENVVAMSNLSEAVQDADLLVFVIPHQFIHRICDEI--TGRVPKKALGITLIK 122
Query 61 GIGVR-NGWPLT---LTHEIGSILDLPKPCILSGANVANDVAAENFAEATIGHPDGETET 116
GI G L + ++G +D+ +L GAN+AN+VAAE F E TIG E
Sbjct 123 GIDEGPEGLKLISDIIREKMG--IDIS---VLMGANIANEVAAEKFCETTIGSK--VMEN 175
Query 117 AALWQLLFDRPNFKINVLPDVDGVEVCGALKNVVALAGGFCEGMGQGTNTKAAILRLGVE 176
L++ L PNF+I V+ D D VE+CGALKN+VA+ GFC+G+ G NTKAA++RLG+
Sbjct 176 GLLFKELLQTPNFRITVVDDADTVELCGALKNIVAVGAGFCDGLRCGDNTKAAVIRLGLM 235
Query 177 EIKTFTMLFF-GDLLADTLYDSAGYADVITTVFGGRNARVAAAFVRQKGKDWDQLEKEML 235
E+ F +F G + T +S G AD+ITT +GGRN RVA AF R GK ++LEKEML
Sbjct 236 EMIAFARIFCKGQVSTATFLESCGVADLITTCYGGRNRRVAEAFART-GKTIEELEKEML 294
Query 236 NGQKLQGPATLELVAEVIRANEVEHLFPLFTSTYAVAFQNADPQAILDVFRTN 288
NGQKLQGP T V +++ + FPLFT+ Y + +++ Q +L +++
Sbjct 295 NGQKLQGPQTSAEVYRILKQKGLLDKFPLFTAVYQICYESRPVQEMLSCLQSH 347
> Hs11440382
Length=349
Score = 233 bits (593), Expect = 6e-61, Method: Compositional matrix adjust.
Identities = 131/277 (47%), Positives = 177/277 (63%), Gaps = 9/277 (3%)
Query 1 YLPGFELPSNLRAVPSLTDACRDADLLVFVMPHQFIERACRELRKAKLLPPHCRAISLIK 60
YLPG +LP N+ AVP + A DAD+L+FV+PHQFI + C +L+ L + ISLIK
Sbjct 63 YLPGHKLPPNVVAVPDVVQAAEDADILIFVVPHQFIGKICDQLKGH--LKANATGISLIK 120
Query 61 GIGVR-NGWPLTLTHEIGSILDLPKPCILSGANVANDVAAENFAEATIGHPDGETETAAL 119
G+ NG L ++ IG L +P +L GAN+A++VA E F E TIG D L
Sbjct 121 GVDEGPNGLKL-ISEVIGERLGIPMS-VLMGANIASEVADEKFCETTIGCKD--PAQGQL 176
Query 120 WQLLFDRPNFKINVLPDVDGVEVCGALKNVVALAGGFCEGMGQGTNTKAAILRLGVEEIK 179
+ L PNF+I V+ +VD VE+CGALKNVVA+ GFC+G+G G NTKAA++RLG+ E+
Sbjct 177 LKELMQTPNFRITVVQEVDTVEICGALKNVVAVGAGFCDGLGFGDNTKAAVIRLGLMEMI 236
Query 180 TFTMLFF-GDLLADTLYDSAGYADVITTVFGGRNARVAAAFVRQKGKDWDQLEKEMLNGQ 238
F LF G + + T +S G AD+ITT +GGRN +VA AF R GK +QLEKE+LNGQ
Sbjct 237 AFAKLFCSGPVSSATFLESCGVADLITTCYGGRNRKVAEAFAR-TGKSIEQLEKELLNGQ 295
Query 239 KLQGPATLELVAEVIRANEVEHLFPLFTSTYAVAFQN 275
KLQGP T + +++ + FPLF + Y V ++
Sbjct 296 KLQGPETARELYSILQHKGLVDKFPLFMAVYKVCYEG 332
> Hs4885291
Length=349
Score = 231 bits (590), Expect = 2e-60, Method: Compositional matrix adjust.
Identities = 131/277 (47%), Positives = 177/277 (63%), Gaps = 9/277 (3%)
Query 1 YLPGFELPSNLRAVPSLTDACRDADLLVFVMPHQFIERACRELRKAKLLPPHCRAISLIK 60
YLPG +LP N+ AVP + A DAD+L+FV+PHQFI + C +L+ L + ISLIK
Sbjct 63 YLPGHKLPPNVVAVPDVVQAAEDADILIFVVPHQFIGKICDQLKGH--LKANPTGISLIK 120
Query 61 GIGVR-NGWPLTLTHEIGSILDLPKPCILSGANVANDVAAENFAEATIGHPDGETETAAL 119
G+ NG L ++ IG L +P +L GAN+A++VA E F E TIG D L
Sbjct 121 GVDEGPNGLKL-ISEVIGERLGIPMS-VLMGANIASEVADEKFCETTIGCKD--PAQGQL 176
Query 120 WQLLFDRPNFKINVLPDVDGVEVCGALKNVVALAGGFCEGMGQGTNTKAAILRLGVEEIK 179
+ L PNF+I V+ +VD VE+CGALKNVVA+ GFC+G+G G NTKAA++RLG+ E+
Sbjct 177 LKELMQTPNFRITVVQEVDTVEICGALKNVVAVGAGFCDGLGFGDNTKAAVIRLGLMEMI 236
Query 180 TFTMLFF-GDLLADTLYDSAGYADVITTVFGGRNARVAAAFVRQKGKDWDQLEKEMLNGQ 238
F LF G + + T +S G AD+ITT +GGRN +VA AF R GK +QLEKE+LNGQ
Sbjct 237 AFAKLFCSGPVSSATFLESCGVADLITTCYGGRNRKVAEAFAR-TGKSIEQLEKELLNGQ 295
Query 239 KLQGPATLELVAEVIRANEVEHLFPLFTSTYAVAFQN 275
KLQGP T + +++ + FPLF + Y V ++
Sbjct 296 KLQGPETARELYSILQHKGLVDKFPLFMAVYKVCYEG 332
> CE00505
Length=351
Score = 224 bits (572), Expect = 2e-58, Method: Compositional matrix adjust.
Identities = 128/295 (43%), Positives = 181/295 (61%), Gaps = 9/295 (3%)
Query 1 YLPGFELPSNLRAVPSLTDACRDADLLVFVMPHQFIERACRELRKAKLLPPHCRAISLIK 60
YLPG LP+N+ AV L ++C +++LVFV+PHQF++ C +L +P +AISLIK
Sbjct 64 YLPGKVLPNNVVAVTDLVESCEGSNVLVFVVPHQFVKGICEKL--VGKIPADTQAISLIK 121
Query 61 GIGV--RNGWPLTLTHEIGSILDLPKPCILSGANVANDVAAENFAEATIGHPDGETETAA 118
G+ R G L ++ EI IL + + +L GAN+A +VA +NF EATIG + E
Sbjct 122 GVSTEKRGGLKL-ISEEIKEILKI-EVSVLMGANLAPEVANDNFCEATIGCKR-KAEDGP 178
Query 119 LWQLLFDRPNFKINVLPDVDGVEVCGALKNVVALAGGFCEGMGQGTNTKAAILRLGVEEI 178
L + LF NF+INV+ D VE+CGALKNVVA A GF +G+G G NTKAA++RLG+ E
Sbjct 179 LLKKLFHTDNFRINVVEDAHTVELCGALKNVVACAAGFTDGLGYGDNTKAAVIRLGLMET 238
Query 179 KTFTMLFFGDLLADTLYDSAGYADVITTVFGGRNARVAAAFVRQKGKDWDQLEKEMLNGQ 238
F ++ T ++S G AD+ITT +GGRN +V AFV+ GK ++EKE+LNGQ
Sbjct 239 TKFVEHYYPGSNLQTFFESCGIADLITTCYGGRNRKVCEAFVKT-GKSMAEVEKELLNGQ 297
Query 239 KLQGPATLELVAEVIRANEVEHLFPLFTSTYAVAFQNADPQAILDVFRTNRPRSI 293
QGP T E V ++ ++ FPLFT+ + + P ++D R N P +
Sbjct 298 SAQGPLTAEEVYLMMHKTGLDAKFPLFTAVHKICAGEMKPAELVDCLR-NHPEHM 351
> 7297034
Length=360
Score = 217 bits (552), Expect = 4e-56, Method: Compositional matrix adjust.
Identities = 132/297 (44%), Positives = 182/297 (61%), Gaps = 8/297 (2%)
Query 1 YLPGFELPSNLRAVPSLTDACRDADLLVFVMPHQFIERACRELRKAKLLPPHCRAISLIK 60
YL G +LP N+ AVP L +A ++AD+L+FV+PHQFI C++L + P+ AISLIK
Sbjct 64 YLKGHKLPPNVVAVPDLVEAAKNADILIFVVPHQFIPNFCKQLLGK--IKPNAIAISLIK 121
Query 61 GIGVRNGWPLTLTHEIGSILDLPKPC-ILSGANVANDVAAENFAEATIGHPDGETETAAL 119
G G + L I + L PC +L GAN+AN+VA NF E TIG D + +
Sbjct 122 GFDKAEGGGIDLISHIIT-RHLKIPCAVLMGANLANEVAEGNFCETTIGCTD--KKYGKV 178
Query 120 WQLLFDRPNFKINVLPDVDGVEVCGALKNVVALAGGFCEGMGQGTNTKAAILRLGVEEIK 179
+ LF +F++ V+ D D VEVCGALKN+VA GF +G+ G NTKAA++RLG+ E+
Sbjct 179 LRDLFQANHFRVVVVDDADAVEVCGALKNIVACGAGFVDGLKLGDNTKAAVIRLGLMEMI 238
Query 180 TFTMLFFGDLLADTLYDSAGYADVITTVFGGRNARVAAAFVRQKGKDWDQLEKEMLNGQK 239
F +F+ T ++S G AD+ITT +GGRN RV+ AFV GK ++LEKEMLNGQK
Sbjct 239 RFVDVFYPGSKLSTFFESCGVADLITTCYGGRNRRVSEAFVTS-GKTIEELEKEMLNGQK 297
Query 240 LQGPATLELVAEVIRANEVEHLFPLFTSTYAVAFQNADPQAILDVFRTNRPRSIQRS 296
LQGP T E V +++ +E FPLFT+ + + P ++D R N P + S
Sbjct 298 LQGPPTAEEVNYMLKNKGLEDKFPLFTAIHKICTNQLKPNDLIDCIR-NHPEHMDTS 353
> SPBC215.05
Length=385
Score = 214 bits (544), Expect = 3e-55, Method: Compositional matrix adjust.
Identities = 127/289 (43%), Positives = 174/289 (60%), Gaps = 8/289 (2%)
Query 1 YLPGFELPSNLRAVPSLTDACRDADLLVFVMPHQFIERACRELRKAKLLPPHCRAISLIK 60
YLPG E P N+ AVP + + R AD+LVFV+PHQFIER C ++ L+ P IS IK
Sbjct 87 YLPGIECPPNVIAVPDVREVARRADILVFVVPHQFIERVCDQM--VGLIRPGAVGISCIK 144
Query 61 GIGVRNGWPLTLTHEIGSILDLPKPC-ILSGANVANDVAAENFAEATIG-HPDGETET-A 117
G+ V + I L + C +LSGANVAN+VA E F E TIG +P E +
Sbjct 145 GVAVSKEGVRLYSEVISEKLGIY--CGVLSGANVANEVAREQFCETTIGFNPPNEVDIPR 202
Query 118 ALWQLLFDRPNFKINVLPDVDGVEVCGALKNVVALAGGFCEGMGQGTNTKAAILRLGVEE 177
+FDRP F + + DV GV + GALKNVVA+A GF +G+ G NTKAAI+R G+ E
Sbjct 203 EQIAAVFDRPYFSVVSVDDVAGVALGGALKNVVAMAVGFADGLEWGGNTKAAIMRRGLLE 262
Query 178 IKTFTMLFFGDLLADTLYDSAGYADVITTVFGGRNARVAAAFVRQKGKDWDQLEKEMLNG 237
++ F FF + S G AD++T+ GGRN R A AFV+ GK + LEKE+L G
Sbjct 263 MQKFATTFFDSDPRTMVEQSCGIADLVTSCLGGRNNRCAEAFVKT-GKSLETLEKELLGG 321
Query 238 QKLQGPATLELVAEVIRANEVEHLFPLFTSTYAVAFQNADPQAILDVFR 286
Q LQG AT + V E + ++ FPLFT+ Y +++++ DP+ ++ V +
Sbjct 322 QLLQGAATSKDVHEFLLTKDMVKDFPLFTAVYNISYEDMDPKDLIIVLQ 370
> YOL059w
Length=440
Score = 205 bits (522), Expect = 1e-52, Method: Compositional matrix adjust.
Identities = 120/283 (42%), Positives = 169/283 (59%), Gaps = 12/283 (4%)
Query 1 YLPGFELPSNLRAVPSLTDACRDADLLVFVMPHQFIERACRELRKAKLLPPHCRAISLIK 60
YLP +LP NL A P L + + AD+LVF +PHQF+ ++L+ + PH RAIS +K
Sbjct 144 YLPNIDLPHNLVADPDLLHSIKGADILVFNIPHQFLPNIVKQLQGH--VAPHVRAISCLK 201
Query 61 GIGVRNGWPLTLTHEIGSILDLPKPCILSGANVANDVAAENFAEATIGHP-------DGE 113
G + + L+ + L + + LSGAN+A +VA E+++E T+ + DG+
Sbjct 202 GFELGSKGVQLLSSYVTDELGI-QCGALSGANLAPEVAKEHWSETTVAYQLPKDYQGDGK 260
Query 114 TETAALWQLLFDRPNFKINVLPDVDGVEVCGALKNVVALAGGFCEGMGQGTNTKAAILRL 173
+ +LLF RP F +NV+ DV G+ + GALKNVVALA GF EGMG G N AAI RL
Sbjct 261 DVDHKILKLLFHRPYFHVNVIDDVAGISIAGALKNVVALACGFVEGMGWGNNASAAIQRL 320
Query 174 GVEEIKTFTMLFFGDLLADTLY-DSAGYADVITTVFGGRNARVAAAFVRQKGKDWDQLEK 232
G+ EI F +FF + +T Y +SAG AD+ITT GGRN +V A ++ + GK + EK
Sbjct 321 GLGEIIKFGRMFFPESKVETYYQESAGVADLITTCSGGRNVKV-ATYMAKTGKSALEAEK 379
Query 233 EMLNGQKLQGPATLELVAEVIRANEVEHLFPLFTSTYAVAFQN 275
E+LNGQ QG T V E ++ E+ FPLF + Y + + N
Sbjct 380 ELLNGQSAQGIITCREVHEWLQTCELTQEFPLFEAVYQIVYNN 422
> CE18701
Length=374
Score = 203 bits (517), Expect = 4e-52, Method: Compositional matrix adjust.
Identities = 121/295 (41%), Positives = 169/295 (57%), Gaps = 11/295 (3%)
Query 1 YLPGFELPSNLRAVPSLTDACRDADLLVFVMPHQFIERACRELRKAKLLPPHCRAISLIK 60
YLPG +P N+ A SL +AC+ A +L+ V+PHQ I + C ELR L AISL K
Sbjct 85 YLPGRRIPDNVVATSSLLEACQSAHILILVVPHQGIPQICDELRGK--LQKGAHAISLTK 142
Query 61 GIG--VRNGWPLT--LTHEIGSILDLPKPCILSGANVANDVAAENFAEATIGHPDGETET 116
GI NG ++ +I L + + +L GAN+A +VA F EATIG +
Sbjct 143 GISSSCENGEIKMQLISEDIERALGV-QCSVLMGANLAGEVADGKFCEATIGCKS--LKN 199
Query 117 AALWQLLFDRPNFKINVLPDVDGVEVCGALKNVVALAGGFCEGMGQGTNTKAAILRLGVE 176
+ +FD PNF+I V D + VE+CGALKN+VA A GF +G+G N K+AI+RLG+
Sbjct 200 GEELKKVFDTPNFRIRVTTDYEAVELCGALKNIVACAAGFADGLGWAYNVKSAIIRLGLL 259
Query 177 EIKTFTMLFFGDLLADTLYDSAGYADVITTVFGGRNARVAAAFVRQKGKDWDQLEKEMLN 236
E K F F+ + T ++S G AD+ITT +GGRN +VA AF++ K +E+E+L
Sbjct 260 ETKKFVEHFYPSSVGHTYFESCGVADLITTCYGGRNRKVAEAFIKSD-KPLRVIEQELLK 318
Query 237 GQKLQGPATLELVAEVIRANEVEHLFPLFTSTYAVAFQNADPQAILDVFRTNRPR 291
GQ QGP T + V E++ NE+ FP+F S + V + Q + D R N P
Sbjct 319 GQSAQGPPTAQDVYEMLEINEISEKFPIFASVHKVFTGHHGEQELYDSLR-NHPE 372
> YDL022w
Length=391
Score = 203 bits (516), Expect = 5e-52, Method: Compositional matrix adjust.
Identities = 119/283 (42%), Positives = 165/283 (58%), Gaps = 12/283 (4%)
Query 1 YLPGFELPSNLRAVPSLTDACRDADLLVFVMPHQFIERACRELRKAKLLPPHCRAISLIK 60
YLPG LP NL A P L D+ +D D++VF +PHQF+ R C +L+ + H RAIS +K
Sbjct 95 YLPGITLPDNLVANPDLIDSVKDVDIIVFNIPHQFLPRICSQLKGH--VDSHVRAISCLK 152
Query 61 GIGVRNGWPLTLTHEIGSILDLPKPCILSGANVANDVAAENFAEATIGHP-------DGE 113
G V L+ I L + + LSGAN+A +VA E+++E T+ + +G+
Sbjct 153 GFEVGAKGVQLLSSYITEELGI-QCGALSGANIATEVAQEHWSETTVAYHIPKDFRGEGK 211
Query 114 TETAALWQLLFDRPNFKINVLPDVDGVEVCGALKNVVALAGGFCEGMGQGTNTKAAILRL 173
+ + LF RP F ++V+ DV G+ +CGALKNVVAL GF EG+G G N AAI R+
Sbjct 212 DVDHKVLKALFHRPYFHVSVIEDVAGISICGALKNVVALGCGFVEGLGWGNNASAAIQRV 271
Query 174 GVEEIKTFTMLFFGDLLADTLY-DSAGYADVITTVFGGRNARVAAAFVRQKGKDWDQLEK 232
G+ EI F +FF + +T Y +SAG AD+ITT GGRN +V A + GKD + EK
Sbjct 272 GLGEIIRFGQMFFPESREETYYQESAGVADLITTCAGGRNVKV-ARLMATSGKDAWECEK 330
Query 233 EMLNGQKLQGPATLELVAEVIRANEVEHLFPLFTSTYAVAFQN 275
E+LNGQ QG T + V E + FPLF + Y + + N
Sbjct 331 ELLNGQSAQGLITCKEVHEWLETCGSVEDFPLFEAVYQIVYNN 373
> SPAC23D3.04c
Length=373
Score = 197 bits (501), Expect = 3e-50, Method: Compositional matrix adjust.
Identities = 116/288 (40%), Positives = 169/288 (58%), Gaps = 10/288 (3%)
Query 1 YLPGFELPSNLRAVPSLTDACRDADLLVFVMPHQFIERACRELRKAKLLPPHCRAISLIK 60
YL G + PSN+ A P + D +D+LV+V+PHQF+ R C +L+ L AIS IK
Sbjct 89 YLKGIKCPSNVFANPDIRDVGSRSDILVWVLPHQFVVRICNQLKGC--LKKDAVAISCIK 146
Query 61 GIGVRNGWPLTLTHEIGSILDLPKPC-ILSGANVANDVAAENFAEATIGH-PDGETE--- 115
G+ V + I + C +LSGAN+A++VA E F E TIG+ P+
Sbjct 147 GVSVTKDRVRLFSDIIEE--NTGMYCGVLSGANIASEVAQEKFCETTIGYLPNSSVNPRY 204
Query 116 TAALWQLLFDRPNFKINVLPDVDGVEVCGALKNVVALAGGFCEGMGQGTNTKAAILRLGV 175
T Q LF+RP F++N++ DV GV + GALKN+VA+A G +G+ G NTK+A++R+G+
Sbjct 205 TPKTIQALFNRPYFRVNIVEDVPGVALGGALKNIVAVAAGIIDGLELGDNTKSAVMRIGL 264
Query 176 EEIKTFTMLFFGDLLADTLYDSAGYADVITTVFGGRNARVAAAFVRQKGKDWDQLEKEML 235
E++ F +FF +S G AD+ITT GGRN + A AFV+ GK +E+E+L
Sbjct 265 LEMQKFGRMFFDCKPLTMSEESCGIADLITTCLGGRNHKCAVAFVKT-GKPMHVVEQELL 323
Query 236 NGQKLQGPATLELVAEVIRANEVEHLFPLFTSTYAVAFQNADPQAILD 283
+GQKLQG AT + V E + FPLFT+ Y + ++ P +L+
Sbjct 324 DGQKLQGAATAKEVYEFLDNQNKVSEFPLFTAVYRIVYEGLPPNKLLE 371
> 7291535
Length=349
Score = 173 bits (438), Expect = 5e-43, Method: Compositional matrix adjust.
Identities = 95/273 (34%), Positives = 151/273 (55%), Gaps = 7/273 (2%)
Query 1 YLPGFELPSNLRAVPSLTDACRDADLLVFVMPHQFIERACRELRKAKLLPPHCRAISLIK 60
Y+P FELP N+ AV + RDAD+++F +P F+ C+ L + P A+SLIK
Sbjct 63 YMPNFELPPNIVAVDDIVTTARDADIIIFAIPPTFVSSCCKTLLGK--VKPTAHAVSLIK 120
Query 61 GIGVRNGWPLTLTHEIGSILDLPKPC-ILSGANVANDVAAENFAEATIGHPDGETETAAL 119
G + L +I + L PC +L G N+A+++A ++FAE T+G D + +
Sbjct 121 GFERGDDGQFVLISQI-IMRQLKIPCSVLVGCNLAHELAHDHFAEGTVGCRD--QKYYRV 177
Query 120 WQLLFDRPNFKINVLPDVDGVEVCGALKNVVALAGGFCEGMGQGTNTKAAILRLGVEEIK 179
+F P F++ V D D VE+C L+N++A A G +GM NTK I+R G E+
Sbjct 178 LHDIFKSPTFRVVVTEDADCVEICSTLRNIIAFAAGCSDGMELNENTKGGIIRRGFLEML 237
Query 180 TFTMLFFGDLLADTLYDSAGYADVITTVFGGRNARVAAAFVRQKGKDWDQLEKEMLNGQK 239
F +F+ T ++S G +D++T+ + RN ++A AFV+ GK +LE ++ G +
Sbjct 238 QFVDVFYPGCRMGTFFESCGISDLVTSCYANRNRKLAEAFVKT-GKPLSELEHILIPGHE 296
Query 240 LQGPATLELVAEVIRANEVEHLFPLFTSTYAVA 272
GP T ELV +++ +E FPLFT+ Y +
Sbjct 297 PLGPVTAELVHHMLKKKGLEDKFPLFTTVYRIC 329
> ECU05g0270
Length=345
Score = 158 bits (399), Expect = 2e-38, Method: Compositional matrix adjust.
Identities = 97/284 (34%), Positives = 158/284 (55%), Gaps = 10/284 (3%)
Query 1 YLPGFELPSNLRAVPSLTDACRDADLLVFVMPHQFIERACRELRKAKLLPPHCRAISLIK 60
YLPG LP NL+AV + + D+D+LVF +PHQ++ A L+ L+ C +SL K
Sbjct 62 YLPGVHLPENLKAVDDIC-SLADSDVLVFALPHQYMG-AIEPLKG--LVKSSCIGVSLTK 117
Query 61 G-IGVRNGWPLTLTHEIGSILDLPKPCILSGANVANDVAAENFAEATIGHPDGETETAAL 119
G + +G ++ I ILD+ ++ GAN+A+ VA + +E T+G+ D + A +
Sbjct 118 GFVSAEDGDIDLVSRLIHRILDINVSVVM-GANIASQVAQDMISEGTLGYTD--EDAADI 174
Query 120 WQLLFDRPNFKINVLPDVDGVEVCGALKNVVALAGGFCEGMGQGTNTKAAILRLGVEEIK 179
LF+ +++ + D+ GVE+ G LKN+V++A GF EG+G TNTK AI R G E++
Sbjct 175 VYKLFNSYAYRVTKIKDIYGVEISGTLKNIVSMAYGFAEGLGYCTNTKVAIFRNGFAEMR 234
Query 180 TFTMLFFGDLLADTLYDSAGYADVITTVFGGRNARVAAAFVRQKGKDWDQLEKEMLNGQK 239
F F+ ++L+ S+G D++ + GRN A + +K + + E+ M K
Sbjct 235 KFFKFFYPMATTESLFQSSGVGDLLVSCMSGRNFG-CARLMAEKRMNLREAEESMCF-TK 292
Query 240 LQGPATLELVAEVIRANEVEHLFPLFTSTYAVAFQNADPQAILD 283
LQGP T +V +R + FPL ++ Y + ++N AIL+
Sbjct 293 LQGPGTALIVYNYLRRQKRVDEFPLMSTVYRICYENEAYDAILE 336
> 7300841
Length=1118
Score = 139 bits (350), Expect = 1e-32, Method: Compositional matrix adjust.
Identities = 92/291 (31%), Positives = 146/291 (50%), Gaps = 27/291 (9%)
Query 1 YLPGFELPSNLRAVPSLTDACRDADLLVFVMPHQFIERACRELRKAKLLPPHCRAISLIK 60
YLPG +LP+NL AV L +A ++AD+LVF P +F++ C L + + A+S+ K
Sbjct 63 YLPGIKLPNNLIAVNDLLEAAQNADILVFSTPLEFVQSYCNIL--SGNVKESAFAVSMTK 120
Query 61 GIGVRNGWPLTL-THEIGSILDLPKPCILSGANVANDVAAENFAEATIGHPDGETETAAL 119
G+ NG + L +H I L +P ++S A+ A ++A E TIG D +
Sbjct 121 GLLSENGEGIELVSHAISESLGIPCYSMMS-AHSAMEMAQGKLCEVTIGCSDNSHSKLLI 179
Query 120 WQLLFDRPNFKINVLPDVDGVEVCGALKNVVALAGGFCEGMGQGTNTKAAILRLGVEEIK 179
+ + N ++ + DVDGVE+CG L +VVAL GF +G+ G N + A + LGV+EI
Sbjct 180 SAMQTN--NCRVISVNDVDGVELCGTLTDVVALGAGFIDGLRLGENARLAAIHLGVKEIM 237
Query 180 TFTMLFFGDLLADTLYDSAGYADVITTVFGGRNARVAAAFVRQKGKDWDQLEKEMLNGQK 239
F FF T Y+S G + + + F ++E + +G+K
Sbjct 238 RFIKTFFPSSKMSTFYESCGVTNAVASSF--------------------EIEANLHSGRK 277
Query 240 LQGPATLELVAEVIRANEVEHLFPLFTSTYAVAFQNADPQAILDVFRTNRP 290
L GP V + ++H FPLFT+ + + A P+ +++ R N P
Sbjct 278 LLGPMVASNVNAFLENGLMQHEFPLFTAIHLICQSEAPPELMIEALR-NHP 327
> At2g40690
Length=433
Score = 59.7 bits (143), Expect = 9e-09, Method: Compositional matrix adjust.
Identities = 76/289 (26%), Positives = 120/289 (41%), Gaps = 28/289 (9%)
Query 1 YLPGFELPSNLRAVPSLTDACRDADLLVFVMPHQFIERACRELRKAKLLPPHCRAISLIK 60
Y P +LP N+ A A DAD + +P QF + A + P ISL K
Sbjct 145 YFPEHKLPENVIATTDAKAALLDADYCLHAVPVQFSSSFLEGI--ADYVDPGLPFISLSK 202
Query 61 GIGVRNGWPLTLTHEIGSI-LDLPK-PCI-LSGANVANDVAAENFAEATIGHPDGETETA 117
G+ + L + +I I L P+ P + LSG + A ++ N A + +
Sbjct 203 GLELNT---LRMMSQIIPIALKNPRQPFVALSGPSFALELM-NNLPTAMVVASKDKKLAN 258
Query 118 ALWQLLFDRPNFKINVLPDVDGVEVCGALKNVVALAGGFCEGMGQGTNTKAAILRLGVEE 177
A+ QLL VE+ GALKNV+A+A G +GM G N+ AA++ G E
Sbjct 259 AVQQLL-------------ASSVEIAGALKNVLAIAAGIVDGMNLGNNSMAALVSQGCSE 305
Query 178 IKTFTMLFFGDLLADTLYDSAGYADVITTVFGGRNARVAAAFVRQKGKDWDQLEKEMLNG 237
I+ G T+ +G D++ T F + G+ D + M
Sbjct 306 IRWLATKVMGA-KPTTITGLSGTGDIMLTCFVNLSRNRTVGVRLGSGETLDDILTSM--N 362
Query 238 QKLQGPATLELVAEVIRANEVEHLFPLFTSTYAVAFQNADP-QAILDVF 285
Q +G AT V + + V+ P+ T+ + P +A+L++
Sbjct 363 QVAEGVATAGAVIALAQKYNVK--LPVLTAVAKIIDNELTPTKAVLELM 409
> At2g41540
Length=462
Score = 47.4 bits (111), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 58/218 (26%), Positives = 89/218 (40%), Gaps = 29/218 (13%)
Query 11 LRAVPSLTDACRDADLLVFVMPHQFIERACRELRK---AKLLPPHCRAISLIKGIGV--- 64
L+ V +L +A DAD++V +P E+ K ++ P ISL KGI
Sbjct 146 LKVVTNLQEAVWDADIVVNGLPSTETREVFEEISKYWKERITVPII--ISLSKGIETALE 203
Query 65 ---RNGWPLTLTHEIGSILDLPKPCILSGANVANDVAAENFAEATIGHPDGETETAALWQ 121
P + H+ + + L G N+A ++ + +A A I AA W+
Sbjct 204 PVPHIITPTKMIHQATGV-PIDNVLYLGGPNIAAEIYNKEYANARIC-------GAAKWR 255
Query 122 L----LFDRPNFKINVLPDVDGVEVCGALKNVVALAGGFCEGMGQGTNTKAAILRLGVEE 177
+P+F + D+ EV G LKNV A+ G + + T ++
Sbjct 256 KPLAKFLRQPHFIVWDNSDLVTHEVMGGLKNVYAIGAGMVAALTNESATSKSVYFAHC-- 313
Query 178 IKTFTMLFFGDLLADTLYDSAG--YADVITTVFGGRNA 213
T M+F LLA+ AG AD T+ GRNA
Sbjct 314 --TSEMIFITHLLAEEPEKLAGPLLADTYVTLLKGRNA 349
> CE15419
Length=441
Score = 32.7 bits (73), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 18/47 (38%), Positives = 25/47 (53%), Gaps = 1/47 (2%)
Query 180 TFTMLFFGDLLADTLYDSAGYADVITTVFGGRNARVAAAFVRQKGKD 226
TF +F D D +Y +A +D I +VF G +A V A + GKD
Sbjct 56 TFDHIFRTDATQDDMY-TAFLSDTINSVFAGNDATVLAMGAKTNGKD 101
> 7290824
Length=1047
Score = 32.3 bits (72), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 19/63 (30%), Positives = 32/63 (50%), Gaps = 5/63 (7%)
Query 69 PLTLTHEI--GSILDL---PKPCILSGANVANDVAAENFAEATIGHPDGETETAALWQLL 123
P+ + HE+ SILDL P+ C+L +V +A F E +G E E A+ + +
Sbjct 316 PMVVIHELFNASILDLTWGPQECLLMACSVDGSIACLKFTEEELGKAISEEEQNAIIRKM 375
Query 124 FDR 126
+ +
Sbjct 376 YGK 378
> At1g12000
Length=566
Score = 32.0 bits (71), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 33/105 (31%), Positives = 45/105 (42%), Gaps = 12/105 (11%)
Query 82 LPKPCILSGANVANDVAAENFAEATIGHPDGETETAALWQLLFDRPNFKINVLPDVDGVE 141
LP P +L GA E A + G+PD E A L+ L+ +P+ + V+PD D
Sbjct 41 LPLPSVLKGAFKI----VEGPASSAAGNPD---EIAKLFPGLYGQPS--VAVVPDQDAPS 91
Query 142 VCGALKNVVALAGGFCEGMGQGTNTKAAILRLGVEEIKTFTMLFF 186
LK V L+GG G G N + + E K T F
Sbjct 92 SAPKLKIGVVLSGGQAPG---GHNVISGLFDYLQERAKGSTFYGF 133
Lambda K H
0.320 0.136 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 8594425070
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40