bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
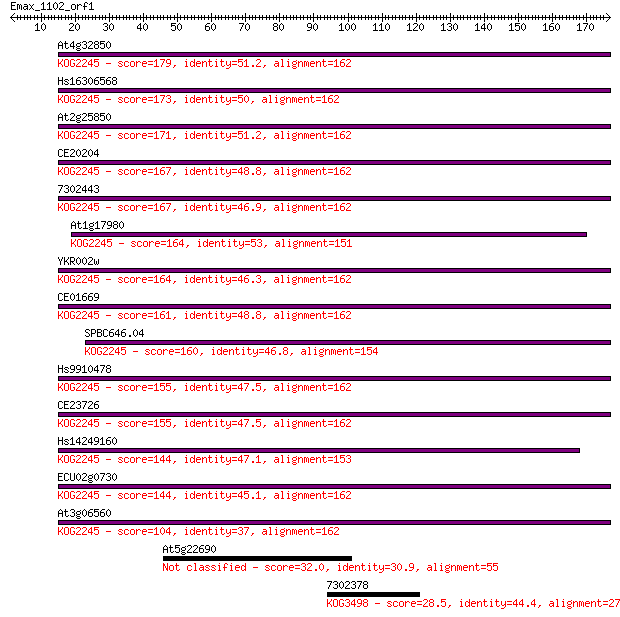
Query= Emax_1102_orf1
Length=176
Score E
Sequences producing significant alignments: (Bits) Value
At4g32850 179 2e-45
Hs16306568 173 2e-43
At2g25850 171 6e-43
CE20204 167 1e-41
7302443 167 1e-41
At1g17980 164 7e-41
YKR002w 164 9e-41
CE01669 161 7e-40
SPBC646.04 160 1e-39
Hs9910478 155 3e-38
CE23726 155 4e-38
Hs14249160 144 7e-35
ECU02g0730 144 1e-34
At3g06560 104 9e-23
At5g22690 32.0 0.71
7302378 28.5 7.2
> At4g32850
Length=729
Score = 179 bits (454), Expect = 2e-45, Method: Composition-based stats.
Identities = 83/162 (51%), Positives = 112/162 (69%), Gaps = 3/162 (1%)
Query 15 FFLLFFLKLQQDNRIKKITPVPDAYTPVLKFTFSGVDIDLLFARLYLRCIPTSWIGLYKD 74
FF++ L + + ++ PVPDA+ PV+KF F G+ IDLL+A + L +P
Sbjct 130 FFIILHDILAEMEEVTELHPVPDAHVPVMKFKFQGIPIDLLYASISLLVVPQDLD--ISS 187
Query 75 NDILRHTDEKTARSLNGSRVADMLLTLVPNKLVFKTSLRFIKQWAKSRGIYSNVMGYLGG 134
+ +L DE T RSLNG RVAD +L LVPN F+T+LR +K WAK RG+YSNV G+LGG
Sbjct 188 SSVLCEVDEPTVRSLNGCRVADQILKLVPNFEHFRTTLRCLKYWAKKRGVYSNVTGFLGG 247
Query 135 ISWAILVARCCQLYPNFPPSLIIQRFFTVYTQWQWSRSPVVL 176
++WA+LVAR CQLYPN PS+++ RFF VYTQW+W +PV+L
Sbjct 248 VNWALLVARVCQLYPNAIPSMLVSRFFRVYTQWRWP-NPVML 288
> Hs16306568
Length=736
Score = 173 bits (439), Expect = 2e-43, Method: Composition-based stats.
Identities = 81/162 (50%), Positives = 114/162 (70%), Gaps = 3/162 (1%)
Query 15 FFLLFFLKLQQDNRIKKITPVPDAYTPVLKFTFSGVDIDLLFARLYLRCIPTSWIGLYKD 74
FF FF KL+ + I+ + V DA+ PV+KF F G++IDL+FARL ++ I + +D
Sbjct 128 FFQSFFEKLKHQDGIRNLRAVEDAFVPVIKFEFDGIEIDLVFARLAIQTISDNLD--LRD 185
Query 75 NDILRHTDEKTARSLNGSRVADMLLTLVPNKLVFKTSLRFIKQWAKSRGIYSNVMGYLGG 134
+ LR D + RSLNG RV D +L LVPNK F+ +LR +K WAK RGIYSN++G+LGG
Sbjct 186 DSRLRSLDIRCIRSLNGCRVTDEILHLVPNKETFRLTLRAVKLWAKRRGIYSNMLGFLGG 245
Query 135 ISWAILVARCCQLYPNFPPSLIIQRFFTVYTQWQWSRSPVVL 176
+SWA+LVAR CQLYPN S ++ +FF V+++W+W +PV+L
Sbjct 246 VSWAMLVARTCQLYPNAAASTLVHKFFLVFSKWEWP-NPVLL 286
> At2g25850
Length=414
Score = 171 bits (433), Expect = 6e-43, Method: Compositional matrix adjust.
Identities = 83/162 (51%), Positives = 112/162 (69%), Gaps = 3/162 (1%)
Query 15 FFLLFFLKLQQDNRIKKITPVPDAYTPVLKFTFSGVDIDLLFARLYLRCIPTSWIGLYKD 74
FF+ F L + + ++ PV DA+ PV+KF F G+ IDLL+A + L IP +
Sbjct 132 FFIFFRDILAEMEEVTELQPVTDAHVPVMKFKFQGISIDLLYASISLLVIPQDLD--ISN 189
Query 75 NDILRHTDEKTARSLNGSRVADMLLTLVPNKLVFKTSLRFIKQWAKSRGIYSNVMGYLGG 134
+ +L DE+T RSLNG RVAD +L LVPN F+T+LR +K WAK RG+YSNV G+LGG
Sbjct 190 SSVLCDVDEQTVRSLNGCRVADQILKLVPNSEHFRTTLRCLKYWAKKRGVYSNVTGFLGG 249
Query 135 ISWAILVARCCQLYPNFPPSLIIQRFFTVYTQWQWSRSPVVL 176
++WA+LVAR CQ YPN PS+++ RFF VYTQW+W +PV+L
Sbjct 250 VNWALLVARLCQFYPNAIPSMLVSRFFRVYTQWRWP-NPVML 290
> CE20204
Length=655
Score = 167 bits (422), Expect = 1e-41, Method: Composition-based stats.
Identities = 79/162 (48%), Positives = 110/162 (67%), Gaps = 3/162 (1%)
Query 15 FFLLFFLKLQQDNRIKKITPVPDAYTPVLKFTFSGVDIDLLFARLYLRCIPTSWIGLYKD 74
FF F L D + ++ V +A+ PV+K +SGV++D+LFARL L+ +P + D
Sbjct 120 FFTSFKEMLNNDPNVTELHGVEEAFVPVMKLKYSGVELDILFARLALKEVPDT--QELSD 177
Query 75 NDILRHTDEKTARSLNGSRVADMLLTLVPNKLVFKTSLRFIKQWAKSRGIYSNVMGYLGG 134
+++LR+ D+++ RSLNG RVA+ LL LVP + F +LR IK WAK+ GIYSN MG+ GG
Sbjct 178 DNLLRNLDQESVRSLNGCRVAEQLLKLVPRQKEFCVTLRAIKLWAKNHGIYSNSMGFFGG 237
Query 135 ISWAILVARCCQLYPNFPPSLIIQRFFTVYTQWQWSRSPVVL 176
I+WAILVAR CQLYPN PS ++ R F +++ W W PVVL
Sbjct 238 ITWAILVARACQLYPNASPSRLVHRMFFIFSTWTWPH-PVVL 278
> 7302443
Length=830
Score = 167 bits (422), Expect = 1e-41, Method: Composition-based stats.
Identities = 76/162 (46%), Positives = 112/162 (69%), Gaps = 3/162 (1%)
Query 15 FFLLFFLKLQQDNRIKKITPVPDAYTPVLKFTFSGVDIDLLFARLYLRCIPTSWIGLYKD 74
+F FF L++ + + V +A+ PV+K F G++IDLLFARL L+ IP + +D
Sbjct 137 YFQSFFEVLKKQPEVTECRSVEEAFVPVIKMNFDGIEIDLLFARLSLKEIPDDFD--LRD 194
Query 75 NDILRHTDEKTARSLNGSRVADMLLTLVPNKLVFKTSLRFIKQWAKSRGIYSNVMGYLGG 134
+++LR+ D ++ RSLNG RV D +L LVPN F+ +LR IK WAK GIYSN +GY GG
Sbjct 195 DNLLRNLDHRSVRSLNGCRVTDEILALVPNIENFRLALRTIKLWAKKHGIYSNSLGYFGG 254
Query 135 ISWAILVARCCQLYPNFPPSLIIQRFFTVYTQWQWSRSPVVL 176
++WA+LVAR CQLYPN + ++ +FF V+++W+W +PV+L
Sbjct 255 VTWAMLVARTCQLYPNAAAATLVHKFFLVFSRWKWP-NPVLL 295
> At1g17980
Length=714
Score = 164 bits (416), Expect = 7e-41, Method: Composition-based stats.
Identities = 80/156 (51%), Positives = 112/156 (71%), Gaps = 7/156 (4%)
Query 19 FFLKLQ----QDNRIKKITPVPDAYTPVLKFTFSGVDIDLLFARLYLRCIPTSWIGLYKD 74
FF +LQ + + ++ PVPDA+ P++ F +GV IDLL+A+L L IP + L +D
Sbjct 121 FFGELQRMLSEMPEVTELHPVPDAHVPLMGFKLNGVSIDLLYAQLPLWVIPED-LDLSQD 179
Query 75 NDILRHTDEKTARSLNGSRVADMLLTLVPN-KLVFKTSLRFIKQWAKSRGIYSNVMGYLG 133
+ IL++ DE+T RSLNG RV D +L LVPN + F+T+LR ++ WAK RG+YSNV G+LG
Sbjct 180 S-ILQNADEQTVRSLNGCRVTDQILRLVPNIQQNFRTTLRCMRFWAKRRGVYSNVSGFLG 238
Query 134 GISWAILVARCCQLYPNFPPSLIIQRFFTVYTQWQW 169
GI+WA+LVAR CQLYPN P++++ RFF V+ QW W
Sbjct 239 GINWALLVARICQLYPNALPNILVSRFFRVFYQWNW 274
> YKR002w
Length=568
Score = 164 bits (415), Expect = 9e-41, Method: Composition-based stats.
Identities = 75/162 (46%), Positives = 110/162 (67%), Gaps = 3/162 (1%)
Query 15 FFLLFFLKLQQDNRIKKITPVPDAYTPVLKFTFSGVDIDLLFARLYLRCIPTSWIGLYKD 74
FF +F L++ + +I PVPDA+ P++K FSG+ IDL+ ARL +P S D
Sbjct 116 FFTVFDSLLRERKELDEIAPVPDAFVPIIKIKFSGISIDLICARLDQPQVPLSLT--LSD 173
Query 75 NDILRHTDEKTARSLNGSRVADMLLTLVPNKLVFKTSLRFIKQWAKSRGIYSNVMGYLGG 134
++LR+ DEK R+LNG+RV D +L LVP VF+ +LR IK WA+ R +Y+N+ G+ GG
Sbjct 174 KNLLRNLDEKDLRALNGTRVTDEILELVPKPNVFRIALRAIKLWAQRRAVYANIFGFPGG 233
Query 135 ISWAILVARCCQLYPNFPPSLIIQRFFTVYTQWQWSRSPVVL 176
++WA+LVAR CQLYPN ++I+ RFF + ++W W + PV+L
Sbjct 234 VAWAMLVARICQLYPNACSAVILNRFFIILSEWNWPQ-PVIL 274
> CE01669
Length=554
Score = 161 bits (407), Expect = 7e-40, Method: Composition-based stats.
Identities = 79/162 (48%), Positives = 111/162 (68%), Gaps = 3/162 (1%)
Query 15 FFLLFFLKLQQDNRIKKITPVPDAYTPVLKFTFSGVDIDLLFARLYLRCIPTSWIGLYKD 74
FF F L ++ + ++ V A+ P++ +SGVDID+LFARL L+ +P L D
Sbjct 122 FFNSFKDILAKNPEVTELCAVEKAFVPIMTLKYSGVDIDILFARLALKSVPEDLNIL--D 179
Query 75 NDILRHTDEKTARSLNGSRVADMLLTLVPNKLVFKTSLRFIKQWAKSRGIYSNVMGYLGG 134
+ +L++ DE++ RSLNG RVA+ LL LVP++ F +LR IK WAK+ GIYSN MG+ GG
Sbjct 180 DSLLKNLDEESVRSLNGCRVAEQLLKLVPHQKNFCITLRAIKLWAKNHGIYSNSMGFFGG 239
Query 135 ISWAILVARCCQLYPNFPPSLIIQRFFTVYTQWQWSRSPVVL 176
I+WAILVAR CQLYPN PS +IQ+ F +++ W W +PV+L
Sbjct 240 ITWAILVARICQLYPNAAPSRLIQKVFFIFSTWNWP-APVLL 280
> SPBC646.04
Length=566
Score = 160 bits (405), Expect = 1e-39, Method: Composition-based stats.
Identities = 72/154 (46%), Positives = 106/154 (68%), Gaps = 3/154 (1%)
Query 23 LQQDNRIKKITPVPDAYTPVLKFTFSGVDIDLLFARLYLRCIPTSWIGLYKDNDILRHTD 82
L++ + + VPDAY P++KF F G+ IDL+FARL + +P DN++L+ +
Sbjct 123 LREREEVTDLAAVPDAYVPIIKFKFLGISIDLIFARLSVPRVPRDL--ELSDNNLLKGVE 180
Query 83 EKTARSLNGSRVADMLLTLVPNKLVFKTSLRFIKQWAKSRGIYSNVMGYLGGISWAILVA 142
E+ SLNG+RV D +L LVPN+ VFK +LR IK WA+ R IY+NV+G+ GG++WA++VA
Sbjct 181 ERCVLSLNGTRVTDQILQLVPNRAVFKHALRAIKFWAQRRAIYANVVGFPGGVAWAMMVA 240
Query 143 RCCQLYPNFPPSLIIQRFFTVYTQWQWSRSPVVL 176
R CQLYPN S+I+ +FF + QW W + P++L
Sbjct 241 RICQLYPNAVSSVIVAKFFRILHQWNWPQ-PILL 273
> Hs9910478
Length=636
Score = 155 bits (393), Expect = 3e-38, Method: Composition-based stats.
Identities = 77/162 (47%), Positives = 112/162 (69%), Gaps = 3/162 (1%)
Query 15 FFLLFFLKLQQDNRIKKITPVPDAYTPVLKFTFSGVDIDLLFARLYLRCIPTSWIGLYKD 74
FF F+ KL+ +K + V +A+ PV+K F G++ID+LFARL L+ IP +D
Sbjct 129 FFTSFYAKLKLQEEVKDLRAVEEAFVPVIKLCFDGIEIDILFARLALQTIPED--LDLRD 186
Query 75 NDILRHTDEKTARSLNGSRVADMLLTLVPNKLVFKTSLRFIKQWAKSRGIYSNVMGYLGG 134
+ +L++ D + RSLNG RV D +L LVPN F+ +LR IK WAK IYSN++G+LGG
Sbjct 187 DSLLKNLDIRCIRSLNGCRVTDEILHLVPNIDNFRLTLRAIKLWAKCHNIYSNILGFLGG 246
Query 135 ISWAILVARCCQLYPNFPPSLIIQRFFTVYTQWQWSRSPVVL 176
+SWA+LVAR CQLYPN S ++++FF V+++W+W +PV+L
Sbjct 247 VSWAMLVARTCQLYPNAVASTLVRKFFLVFSEWEWP-NPVLL 287
> CE23726
Length=554
Score = 155 bits (392), Expect = 4e-38, Method: Composition-based stats.
Identities = 77/162 (47%), Positives = 110/162 (67%), Gaps = 3/162 (1%)
Query 15 FFLLFFLKLQQDNRIKKITPVPDAYTPVLKFTFSGVDIDLLFARLYLRCIPTSWIGLYKD 74
FF F L +D + + V +A+ P++ +SG++ID LFARL L+ +P D
Sbjct 127 FFSSFKQMLAKDPEVTNLCAVENAFVPIMTMKYSGIEIDFLFARLALKEVPED--LDILD 184
Query 75 NDILRHTDEKTARSLNGSRVADMLLTLVPNKLVFKTSLRFIKQWAKSRGIYSNVMGYLGG 134
+ +L++ D+++ RSLNG RVA+ LL LVPN+ F +LR IK WAK+ GIYSN MG+ GG
Sbjct 185 DALLKNLDQESVRSLNGCRVAENLLKLVPNQENFCITLRAIKLWAKNHGIYSNSMGFFGG 244
Query 135 ISWAILVARCCQLYPNFPPSLIIQRFFTVYTQWQWSRSPVVL 176
I+WAILVAR CQLYPN PS +IQ+ F +++ W+W +PV+L
Sbjct 245 ITWAILVARACQLYPNASPSKLIQKVFFIFSTWKWP-APVLL 285
> Hs14249160
Length=285
Score = 144 bits (364), Expect = 7e-35, Method: Compositional matrix adjust.
Identities = 72/153 (47%), Positives = 101/153 (66%), Gaps = 2/153 (1%)
Query 15 FFLLFFLKLQQDNRIKKITPVPDAYTPVLKFTFSGVDIDLLFARLYLRCIPTSWIGLYKD 74
FF F+ KL+ +K + V +A+ PV+K F G++ID+LFARL L+ IP
Sbjct 129 FFTSFYDKLKLQEEVKDLRAVEEAFVPVIKLCFDGIEIDILFARLALQTIPEDLDLRDDS 188
Query 75 NDILRHTDEKTARSLNGSRVADMLLTLVPNKLVFKTSLRFIKQWAKSRGIYSNVMGYLGG 134
+L++ D + RSLNG RV D +L LVPN F+ +LR IK WAK IYSN++G+LGG
Sbjct 189 --LLKNLDIRCIRSLNGCRVTDEILHLVPNIDNFRLTLRAIKLWAKRHNIYSNILGFLGG 246
Query 135 ISWAILVARCCQLYPNFPPSLIIQRFFTVYTQW 167
+SWA+LVAR CQLYPN S ++ +FF V+++W
Sbjct 247 VSWAMLVARTCQLYPNAIASTLVHKFFLVFSKW 279
> ECU02g0730
Length=505
Score = 144 bits (362), Expect = 1e-34, Method: Compositional matrix adjust.
Identities = 73/163 (44%), Positives = 106/163 (65%), Gaps = 5/163 (3%)
Query 15 FFLLFFLKLQQDNRIKKITPVPDAYTPVLKFTFSGVDIDLLFARLYLRCIPTSWIGLYKD 74
FF F+ +L+ D I+++T + DA+ P++K F G+ IDL+FARL IP G+
Sbjct 118 FFTHFYEELKGDPNIEEVTKIEDAFVPIIKLKFQGIPIDLVFARL---SIPVVKDGINLL 174
Query 75 ND-ILRHTDEKTARSLNGSRVADMLLTLVPNKLVFKTSLRFIKQWAKSRGIYSNVMGYLG 133
ND +L+ DEK SLNGSRV D +L LVP+ F ++LR IK WAK R +Y N GY G
Sbjct 175 NDTLLKSMDEKCILSLNGSRVTDEMLNLVPSVKTFHSALRCIKYWAKRRCVYGNPYGYFG 234
Query 134 GISWAILVARCCQLYPNFPPSLIIQRFFTVYTQWQWSRSPVVL 176
G+++++ VAR CQLYPN I+ +FF +++ W+W +PV+L
Sbjct 235 GVAFSLCVARVCQLYPNASSFTIVCKFFELFSSWKWP-TPVIL 276
> At3g06560
Length=482
Score = 104 bits (259), Expect = 9e-23, Method: Compositional matrix adjust.
Identities = 60/162 (37%), Positives = 92/162 (56%), Gaps = 3/162 (1%)
Query 15 FFLLFFLKLQQDNRIKKITPVPDAYTPVLKFTFSGVDIDLLFARLYLRCIPTSWIGLYKD 74
FF+ L+ + ++ V DA P+++F F G+ +DL +A+L + IP + L +
Sbjct 84 FFISLRDMLKSRREVSELHCVKDAKVPLIRFKFDGILVDLPYAQLRVLSIPNNVDVL--N 141
Query 75 NDILRHTDEKTARSLNGSRVADMLLTLVPNKLVFKTSLRFIKQWAKSRGIYSNVMGYLGG 134
LR DE + + L+G R +L LVP+ +F++ LR +K WAK RG+Y N+ G+LGG
Sbjct 142 PFFLRDIDETSWKILSGVRANKCILQLVPSLELFQSLLRCVKLWAKRRGVYGNLNGFLGG 201
Query 135 ISWAILVARCCQLYPNFPPSLIIQRFFTVYTQWQWSRSPVVL 176
+ AIL A C PN S ++ FF + WQW +PVVL
Sbjct 202 VHMAILAAFVCGYQPNATLSSLLANFFYTFAHWQWP-TPVVL 242
> At5g22690
Length=1008
Score = 32.0 bits (71), Expect = 0.71, Method: Composition-based stats.
Identities = 17/57 (29%), Positives = 32/57 (56%), Gaps = 2/57 (3%)
Query 46 TFSGVDIDLLFARLYLRCIPTSWIGLYKDNDILRHT--DEKTARSLNGSRVADMLLT 100
+FSG D+ + F +L+ + I ++KDNDI R D + ++ SR+A ++ +
Sbjct 15 SFSGEDVRVTFLSHFLKELDRKLISVFKDNDIQRSQSLDPELKLAIRDSRIAIVVFS 71
> 7302378
Length=105
Score = 28.5 bits (62), Expect = 7.2, Method: Compositional matrix adjust.
Identities = 12/27 (44%), Positives = 19/27 (70%), Gaps = 0/27 (0%)
Query 94 VADMLLTLVPNKLVFKTSLRFIKQWAK 120
+ D+L+ +VP+K +K SLRF K+ K
Sbjct 39 IKDLLILMVPSKDFYKNSLRFYKRCTK 65
Lambda K H
0.330 0.144 0.463
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2707167450
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40