bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_1105_orf1
Length=116
Score E
Sequences producing significant alignments: (Bits) Value
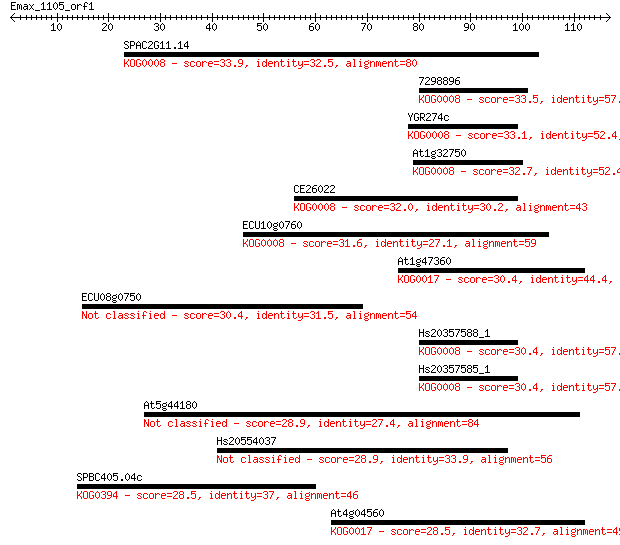
SPAC2G11.14 33.9 0.069
7298896 33.5 0.097
YGR274c 33.1 0.14
At1g32750 32.7 0.16
CE26022 32.0 0.25
ECU10g0760 31.6 0.35
At1g47360 30.4 0.83
ECU08g0750 30.4 0.90
Hs20357588_1 30.4 0.91
Hs20357585_1 30.4 0.91
At5g44180 28.9 2.2
Hs20554037 28.9 2.7
SPBC405.04c 28.5 2.9
At4g04560 28.5 3.4
> SPAC2G11.14
Length=979
Score = 33.9 bits (76), Expect = 0.069, Method: Composition-based stats.
Identities = 26/92 (28%), Positives = 44/92 (47%), Gaps = 12/92 (13%)
Query 23 KRRQQVGDSFAAERCILIYGAENIKAFLAWR-DKRLAK-----KRQRQMNAVHSKEALAG 76
K+R+++ + A ++ G E I R ++ LAK +R+R +A L G
Sbjct 878 KKRREIDEQSTALDAVVPTGDEAIDRRNRRRLEQELAKSQKNWERRRARHAAKEGINLNG 937
Query 77 K------RICRACGRPGHIASNVNCPLYRGPK 102
+ R C CG+ GH+ +N CPL+ P+
Sbjct 938 EGRKPTTRKCSNCGQVGHMKTNKICPLFGRPE 969
> 7298896
Length=2065
Score = 33.5 bits (75), Expect = 0.097, Method: Composition-based stats.
Identities = 12/21 (57%), Positives = 15/21 (71%), Gaps = 0/21 (0%)
Query 80 CRACGRPGHIASNVNCPLYRG 100
C ACG+ GH+ +N CPLY G
Sbjct 1348 CGACGQVGHMRTNKACPLYSG 1368
> YGR274c
Length=1066
Score = 33.1 bits (74), Expect = 0.14, Method: Composition-based stats.
Identities = 11/21 (52%), Positives = 15/21 (71%), Gaps = 0/21 (0%)
Query 78 RICRACGRPGHIASNVNCPLY 98
R C CG+ GHI +N +CP+Y
Sbjct 1036 RRCATCGQIGHIRTNKSCPMY 1056
> At1g32750
Length=1919
Score = 32.7 bits (73), Expect = 0.16, Method: Composition-based stats.
Identities = 11/21 (52%), Positives = 16/21 (76%), Gaps = 0/21 (0%)
Query 79 ICRACGRPGHIASNVNCPLYR 99
+C ACG+ GH+ +N +CP YR
Sbjct 1398 VCGACGQHGHMRTNKHCPRYR 1418
> CE26022
Length=1792
Score = 32.0 bits (71), Expect = 0.25, Method: Composition-based stats.
Identities = 13/43 (30%), Positives = 22/43 (51%), Gaps = 0/43 (0%)
Query 56 RLAKKRQRQMNAVHSKEALAGKRICRACGRPGHIASNVNCPLY 98
++ K ++++ + K C AC GH+ +N NCPLY
Sbjct 1244 KVQKMTEKKVKPIKPPNPNLQKMRCSACHAYGHMKTNRNCPLY 1286
> ECU10g0760
Length=883
Score = 31.6 bits (70), Expect = 0.35, Method: Composition-based stats.
Identities = 16/59 (27%), Positives = 29/59 (49%), Gaps = 13/59 (22%)
Query 46 IKAFLAWRDKRLAKKRQRQMNAVHSKEALAGKRICRACGRPGHIASNVNCPLYRGPKRL 104
IKA+L R K+++++R+ G C CG+ GH+ +N CP + ++
Sbjct 706 IKAYLKARKKKISEERK-------------GVLTCGNCGQVGHMKTNKACPKFASATKM 751
> At1g47360
Length=1182
Score = 30.4 bits (67), Expect = 0.83, Method: Composition-based stats.
Identities = 16/37 (43%), Positives = 20/37 (54%), Gaps = 1/37 (2%)
Query 76 GKRICRACGRPGHIASNVNCPLYRG-PKRLGSAVGET 111
GKR+C CG+ GH L R K+ GS VGE+
Sbjct 207 GKRVCWICGKEGHFKKQCYKWLERNKSKQQGSDVGES 243
> ECU08g0750
Length=212
Score = 30.4 bits (67), Expect = 0.90, Method: Compositional matrix adjust.
Identities = 17/56 (30%), Positives = 26/56 (46%), Gaps = 2/56 (3%)
Query 15 YHVCGGLRKRRQQVGDSFAAERCILIYGAENIKAFLAWRD--KRLAKKRQRQMNAV 68
Y GL +++ VG + R LI N+KA + W R+A KR + N +
Sbjct 73 YPTRDGLSRKKASVGHKMESRREALIEKDSNVKAEMKWEGSRSRVASKRTVERNTI 128
> Hs20357588_1
Length=1689
Score = 30.4 bits (67), Expect = 0.91, Method: Composition-based stats.
Identities = 11/19 (57%), Positives = 13/19 (68%), Gaps = 0/19 (0%)
Query 80 CRACGRPGHIASNVNCPLY 98
C ACG GH+ +N CPLY
Sbjct 1264 CGACGAIGHMRTNKFCPLY 1282
> Hs20357585_1
Length=1710
Score = 30.4 bits (67), Expect = 0.91, Method: Composition-based stats.
Identities = 11/19 (57%), Positives = 13/19 (68%), Gaps = 0/19 (0%)
Query 80 CRACGRPGHIASNVNCPLY 98
C ACG GH+ +N CPLY
Sbjct 1285 CGACGAIGHMRTNKFCPLY 1303
> At5g44180
Length=1694
Score = 28.9 bits (63), Expect = 2.2, Method: Composition-based stats.
Identities = 23/84 (27%), Positives = 35/84 (41%), Gaps = 5/84 (5%)
Query 27 QVGDSFAAERCILIYGAENIKAFLAWRDKRLAKKRQRQMNAVHSKEALAGKRICRACGRP 86
++GDS A ERC ++ + +F W + +K++ RICR C
Sbjct 1183 ELGDSNAVERCSVL---QRFHSFEKWMWDNMLHPSALSAFKYGAKQSSPLFRICRICAEL 1239
Query 87 GHIASNVNCPLYRGPKRLGSAVGE 110
H ++ CP G G VGE
Sbjct 1240 -HFVGDICCPSC-GQMHAGPDVGE 1261
> Hs20554037
Length=129
Score = 28.9 bits (63), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 19/61 (31%), Positives = 28/61 (45%), Gaps = 6/61 (9%)
Query 41 YGAENIKAFLAWRDKRLAKKRQRQMNAVHSKEALAGK-----RICRACGRPGHIASNVNC 95
YG L RD +AKKRQ ++ AV +E LA R+ R R + + ++
Sbjct 30 YGTWKAAEMLG-RDSEMAKKRQTELEAVTFQEGLAESGSRKMRLQRPLSRQAWMWALLDW 88
Query 96 P 96
P
Sbjct 89 P 89
> SPBC405.04c
Length=205
Score = 28.5 bits (62), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 17/51 (33%), Positives = 29/51 (56%), Gaps = 7/51 (13%)
Query 14 LYHVCGGLRKRRQQVGDSF--AAERCILIYGAENIKAFL---AWRDKRLAK 59
L+ G ++R Q +G +F A+ C+L+Y N K+F +WRD+ L +
Sbjct 61 LWDTAG--QERFQSLGVAFYRGADCCVLVYDVNNSKSFETLDSWRDEFLIQ 109
> At4g04560
Length=590
Score = 28.5 bits (62), Expect = 3.4, Method: Composition-based stats.
Identities = 16/49 (32%), Positives = 27/49 (55%), Gaps = 3/49 (6%)
Query 63 RQMNAVHSKEALAGKRICRACGRPGHIASNVNCPLYRGPKRLGSAVGET 111
++ V S++ +R C C +PGH+A NC L R +R+ ++ ET
Sbjct 185 KKNKGVLSQQVGNNERRCFVCNKPGHLAK--NCRLRR-TERVDLSLEET 230
Lambda K H
0.324 0.136 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1174970866
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40