bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_1113_orf1
Length=123
Score E
Sequences producing significant alignments: (Bits) Value
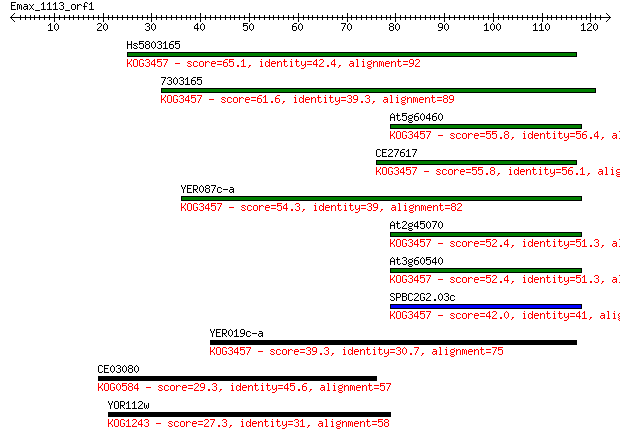
Hs5803165 65.1 3e-11
7303165 61.6 4e-10
At5g60460 55.8 2e-08
CE27617 55.8 2e-08
YER087c-a 54.3 5e-08
At2g45070 52.4 2e-07
At3g60540 52.4 2e-07
SPBC2G2.03c 42.0 3e-04
YER019c-a 39.3 0.002
CE03080 29.3 2.0
YOR112w 27.3 6.8
> Hs5803165
Length=96
Score = 65.1 bits (157), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 39/94 (41%), Positives = 55/94 (58%), Gaps = 8/94 (8%)
Query 25 TMVGTNSLSTQASQTTVGGSRVAAT--RKRVTSKSSTASSGAATISRPRSNAANQGILKF 82
T GTN S+ S + +R A + R+R + T S+G T +A G+ +F
Sbjct 5 TPSGTNVGSSGRSPSKAVAARAAGSTVRQRKNASCGTRSAGRTT------SAGTGGMWRF 58
Query 83 YTEDAPGLRVGPQAVLIMALVFMGVVVMLHIVGK 116
YTED+PGL+VGP VL+M+L+F+ V MLHI GK
Sbjct 59 YTEDSPGLKVGPVPVLVMSLLFIASVFMLHIWGK 92
> 7303165
Length=100
Score = 61.6 bits (148), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 35/103 (33%), Positives = 58/103 (56%), Gaps = 17/103 (16%)
Query 32 LSTQASQTTVG-GSRVAATRKRVTSKSSTASSGAATISRPR-------------SNAANQ 77
+ AS T+VG GSR + ++++ S S G +T+ + + A
Sbjct 1 MPAPASSTSVGSGSR---SPSKLSAPRSAGSGGGSTLKQRKTTTSTTAARSRAPGGAGTG 57
Query 78 GILKFYTEDAPGLRVGPQAVLIMALVFMGVVVMLHIVGKIRQA 120
G+ +FYT+D+PG++VGP VL+M+L+F+ V MLHI GK ++
Sbjct 58 GMWRFYTDDSPGIKVGPVPVLVMSLLFIASVFMLHIWGKYNRS 100
> At5g60460
Length=109
Score = 55.8 bits (133), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 22/39 (56%), Positives = 31/39 (79%), Gaps = 0/39 (0%)
Query 79 ILKFYTEDAPGLRVGPQAVLIMALVFMGVVVMLHIVGKI 117
+L+FYT+DAPGL++ P VLIM+L F+G V LH+ GK+
Sbjct 62 MLRFYTDDAPGLKISPTVVLIMSLCFIGFVTALHVFGKL 100
> CE27617
Length=81
Score = 55.8 bits (133), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 23/41 (56%), Positives = 32/41 (78%), Gaps = 0/41 (0%)
Query 76 NQGILKFYTEDAPGLRVGPQAVLIMALVFMGVVVMLHIVGK 116
N G+ +FYTED+ GL++GP VL+M+LVF+ V +LHI GK
Sbjct 35 NGGLWRFYTEDSTGLKIGPVPVLVMSLVFIASVFVLHIWGK 75
> YER087c-a
Length=82
Score = 54.3 bits (129), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 32/83 (38%), Positives = 48/83 (57%), Gaps = 8/83 (9%)
Query 36 ASQTTVGGSRVAATRKRVTSKSSTASSGAATISRPRSNA-ANQGILKFYTEDAPGLRVGP 94
+S T GG R RK+ +S+ AS+ P+ N +N ILK Y+++A GLRV P
Sbjct 2 SSPTPPGGQRTLQKRKQGSSQKVAASA-------PKKNTNSNNSILKIYSDEATGLRVDP 54
Query 95 QAVLIMALVFMGVVVMLHIVGKI 117
VL +A+ F+ VV LH++ K+
Sbjct 55 LVVLFLAVGFIFSVVALHVISKV 77
> At2g45070
Length=82
Score = 52.4 bits (124), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 20/39 (51%), Positives = 32/39 (82%), Gaps = 0/39 (0%)
Query 79 ILKFYTEDAPGLRVGPQAVLIMALVFMGVVVMLHIVGKI 117
+L+FYT+DAPGL++ P VLIM++ F+ V +LH++GK+
Sbjct 40 MLQFYTDDAPGLKISPNVVLIMSIGFIAFVAVLHVMGKL 78
> At3g60540
Length=81
Score = 52.4 bits (124), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 20/39 (51%), Positives = 32/39 (82%), Gaps = 0/39 (0%)
Query 79 ILKFYTEDAPGLRVGPQAVLIMALVFMGVVVMLHIVGKI 117
+L+FYT+DAPGL++ P VLIM++ F+ V +LH++GK+
Sbjct 37 MLQFYTDDAPGLKISPNVVLIMSIGFIAFVAVLHVMGKL 75
> SPBC2G2.03c
Length=102
Score = 42.0 bits (97), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 16/39 (41%), Positives = 28/39 (71%), Gaps = 0/39 (0%)
Query 79 ILKFYTEDAPGLRVGPQAVLIMALVFMGVVVMLHIVGKI 117
+LK YT++A G +V P V+++++ F+ V +LHIV +I
Sbjct 57 LLKLYTDEASGFKVDPVVVMVLSVGFIASVFLLHIVARI 95
> YER019c-a
Length=88
Score = 39.3 bits (90), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 23/76 (30%), Positives = 39/76 (51%), Gaps = 1/76 (1%)
Query 42 GGSRVAATRKRVTS-KSSTASSGAATISRPRSNAANQGILKFYTEDAPGLRVGPQAVLIM 100
GG R+ R++ S K A + + ++ ILK YT++A G RV VL +
Sbjct 8 GGQRILQKRRQAQSIKEKQAKQTPTSTRQAGYGGSSSSILKLYTDEANGFRVDSLVVLFL 67
Query 101 ALVFMGVVVMLHIVGK 116
++ F+ V+ LH++ K
Sbjct 68 SVGFIFSVIALHLLTK 83
> CE03080
Length=1851
Score = 29.3 bits (64), Expect = 2.0, Method: Composition-based stats.
Identities = 26/67 (38%), Positives = 33/67 (49%), Gaps = 11/67 (16%)
Query 19 SFTLLFTMVGTNSLSTQAS----------QTTVGGSRVAATRKRVTSKSSTASSGAATIS 68
+ T L + +G SLS AS TTV S V ATR + S+SST SS + S
Sbjct 1718 TLTALQSALGNASLSLPASPPPNTETTKVNTTVIPSDVLATRMTM-SQSSTKSSNVSVSS 1776
Query 69 RPRSNAA 75
R R N +
Sbjct 1777 RHRDNQS 1783
> YOR112w
Length=761
Score = 27.3 bits (59), Expect = 6.8, Method: Composition-based stats.
Identities = 18/60 (30%), Positives = 32/60 (53%), Gaps = 2/60 (3%)
Query 21 TLLFTMVGTNSLSTQASQTTVGGSRVAATRKRVTSKSSTASSGAAT--ISRPRSNAANQG 78
TL ++ ++ LS + +TT+ R A+ VT+KSS ++ A + IS R + +G
Sbjct 684 TLAKSIAPSSRLSIKKKKTTILAPRNIASNSTVTTKSSLSNKTARSKPISSIRGSVTKKG 743
Lambda K H
0.324 0.131 0.361
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1191192512
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40