bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
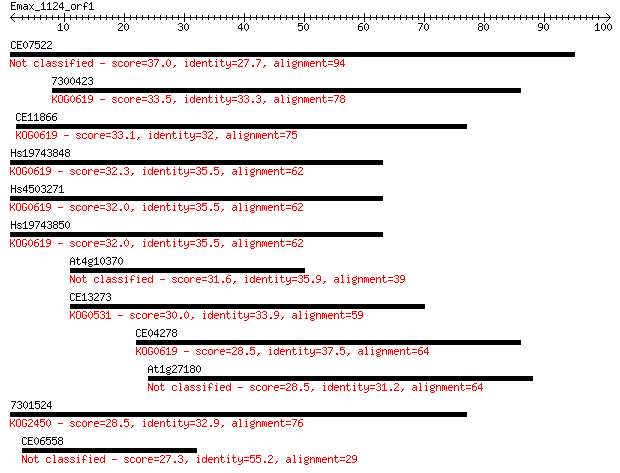
Query= Emax_1124_orf1
Length=100
Score E
Sequences producing significant alignments: (Bits) Value
CE07522 37.0 0.008
7300423 33.5 0.11
CE11866 33.1 0.14
Hs19743848 32.3 0.24
Hs4503271 32.0 0.26
Hs19743850 32.0 0.28
At4g10370 31.6 0.33
CE13273 30.0 1.1
CE04278 28.5 3.0
At1g27180 28.5 3.3
7301524 28.5 3.6
CE06558 27.3 6.8
> CE07522
Length=180
Score = 37.0 bits (84), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 26/94 (27%), Positives = 49/94 (52%), Gaps = 10/94 (10%)
Query 1 DLLLSGGGIERLPYPLLLEATNAERLFLSFNCLEDSAMLDWHVPYLPFLRELSVDNTSIS 60
++LSG + +P +L T E+L L N L +++ +++P L+ELSV N I
Sbjct 50 HVVLSGLCLTSIPSTVLKNRTRIEKLTLDNNVLTENS---FNMPKFANLKELSVRNNKIR 106
Query 61 RLSLLLFSVAELPRLERVNAARTKLLPVEDDPAL 94
L +LL ++ + N ++L V+++P +
Sbjct 107 NLGVLLANIQK-------NCPNIEVLRVKNNPGI 133
> 7300423
Length=537
Score = 33.5 bits (75), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 26/86 (30%), Positives = 43/86 (50%), Gaps = 10/86 (11%)
Query 8 GIERLPYPLLLEATNAERLFLSFNCLEDSAMLD-------WHVPYLPF-LRELSVDNTSI 59
G++ L PL ++ N + L++S+N + D A++ + Y P LR+L + + I
Sbjct 117 GLQDLQAPLFMDVPNVQALYISWNDITDDALVPDLFRGPFRNTRYEPIGLRDLDLSHNRI 176
Query 60 SRLSLLLFSVAELPRLERVNAARTKL 85
RL LF P L ++N A KL
Sbjct 177 VRLDRRLFE--HTPHLTKLNLAYNKL 200
> CE11866
Length=961
Score = 33.1 bits (74), Expect = 0.14, Method: Composition-based stats.
Identities = 24/75 (32%), Positives = 41/75 (54%), Gaps = 4/75 (5%)
Query 2 LLLSGGGIERLPYPLLLEATNAERLFLSFNCLEDSAMLDWHVPYLPFLRELSVDNTSISR 61
L +SG ++ +P +L A N L L N + S + ++ + LPFLREL V N ++ R
Sbjct 180 LDVSGNCLDAIPAQILRNAANLMYLDLGSNNI--SEINNFELMNLPFLRELRVQNNTLRR 237
Query 62 LSLLLFSVAELPRLE 76
+ + F +P+L+
Sbjct 238 IHPMAF--MNVPQLQ 250
> Hs19743848
Length=250
Score = 32.3 bits (72), Expect = 0.24, Method: Compositional matrix adjust.
Identities = 22/62 (35%), Positives = 33/62 (53%), Gaps = 2/62 (3%)
Query 1 DLLLSGGGIERLPYPLLLEATNAERLFLSFNCLEDSAMLDWHVPYLPFLRELSVDNTSIS 60
+L L G I R+ L N +L LSFN + SA+ + + P LREL +DN ++
Sbjct 116 ELHLDGNKISRVDAASLKGLNNLAKLGLSFNSI--SAVDNGSLANTPHLRELHLDNNKLT 173
Query 61 RL 62
R+
Sbjct 174 RV 175
> Hs4503271
Length=359
Score = 32.0 bits (71), Expect = 0.26, Method: Composition-based stats.
Identities = 22/62 (35%), Positives = 33/62 (53%), Gaps = 2/62 (3%)
Query 1 DLLLSGGGIERLPYPLLLEATNAERLFLSFNCLEDSAMLDWHVPYLPFLRELSVDNTSIS 60
+L L G I R+ L N +L LSFN + SA+ + + P LREL +DN ++
Sbjct 225 ELHLDGNKISRVDAASLKGLNNLAKLGLSFNSI--SAVDNGSLANTPHLRELHLDNNKLT 282
Query 61 RL 62
R+
Sbjct 283 RV 284
> Hs19743850
Length=212
Score = 32.0 bits (71), Expect = 0.28, Method: Compositional matrix adjust.
Identities = 22/62 (35%), Positives = 33/62 (53%), Gaps = 2/62 (3%)
Query 1 DLLLSGGGIERLPYPLLLEATNAERLFLSFNCLEDSAMLDWHVPYLPFLRELSVDNTSIS 60
+L L G I R+ L N +L LSFN + SA+ + + P LREL +DN ++
Sbjct 78 ELHLDGNKISRVDAASLKGLNNLAKLGLSFNSI--SAVDNGSLANTPHLRELHLDNNKLT 135
Query 61 RL 62
R+
Sbjct 136 RV 137
> At4g10370
Length=676
Score = 31.6 bits (70), Expect = 0.33, Method: Composition-based stats.
Identities = 14/39 (35%), Positives = 22/39 (56%), Gaps = 2/39 (5%)
Query 11 RLPYPLLLEATNAERLFLSFNCLEDSAMLDWHVPYLPFL 49
R P+P+ + N +F S++C+EDSAM + FL
Sbjct 638 RCPFPIFFKGHNT--IFCSWDCVEDSAMRSYQRLLYSFL 674
> CE13273
Length=1152
Score = 30.0 bits (66), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 20/61 (32%), Positives = 28/61 (45%), Gaps = 2/61 (3%)
Query 11 RLPYPLLLEATNAERLFLSFNCLEDSAMLDWHVPYLPF--LRELSVDNTSISRLSLLLFS 68
RL + + N L L+ NCL+ A+ D PF L L + N SIS S+L
Sbjct 866 RLSSVREISSFNITHLILNNNCLKSIAVNDGQTSLQPFPYLENLDISNNSISNTSILRLG 925
Query 69 V 69
+
Sbjct 926 I 926
> CE04278
Length=1066
Score = 28.5 bits (62), Expect = 3.0, Method: Composition-based stats.
Identities = 24/67 (35%), Positives = 34/67 (50%), Gaps = 12/67 (17%)
Query 22 NAERLFLSFNCLEDSAMLDWHVPYL---PFLRELSVDNTSISRLSLLLFSVAELPRLERV 78
N E LFL N L H P L LR L +DN I ++ FS+A+LP+L+ +
Sbjct 369 NLESLFLQNNQLA-------HFPSLFRLDKLRHLMLDNNQIQKIDN--FSLADLPKLQHL 419
Query 79 NAARTKL 85
+ A +L
Sbjct 420 SLAGNQL 426
> At1g27180
Length=1556
Score = 28.5 bits (62), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 20/64 (31%), Positives = 36/64 (56%), Gaps = 2/64 (3%)
Query 24 ERLFLSFNCLEDSAMLDWHVPYLPFLRELSVDNTSISRLSLLLFSVAELPRLERVNAART 83
E+ FLS C + ++L ++ +P L+EL +D T+IS L +F + +L +L +
Sbjct 917 EKFFLS-GC-SNLSVLPENIGSMPCLKELLLDGTAISNLPYSIFRLQKLEKLSLMGCRSI 974
Query 84 KLLP 87
+ LP
Sbjct 975 EELP 978
> 7301524
Length=659
Score = 28.5 bits (62), Expect = 3.6, Method: Composition-based stats.
Identities = 25/77 (32%), Positives = 39/77 (50%), Gaps = 6/77 (7%)
Query 1 DLLLSGGGIERLPYPLLLE-ATNAERLFLSFNCLEDSAMLDWHVPYLPFLRELSVDNTSI 59
+L+L + LP L+ + A + E+L LS NCL+D L W + LR L +D+ +
Sbjct 18 ELILVQQNLRTLPQELVKKHADHVEQLDLSHNCLKD---LSWLADFEQ-LRHLVLDSNRM 73
Query 60 SRLSLLLFSVAELPRLE 76
L + LP+LE
Sbjct 74 HEAHLRTLT-CPLPQLE 89
> CE06558
Length=562
Score = 27.3 bits (59), Expect = 6.8, Method: Composition-based stats.
Identities = 16/31 (51%), Positives = 20/31 (64%), Gaps = 2/31 (6%)
Query 3 LLSGGGIERLPYPLLLEATNA--ERLFLSFN 31
LLSG L +PLLL TN+ RLF++FN
Sbjct 418 LLSGHCSGHLIFPLLLTLTNSSNSRLFIAFN 448
Lambda K H
0.321 0.141 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1187882580
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40