bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_1145_orf1
Length=158
Score E
Sequences producing significant alignments: (Bits) Value
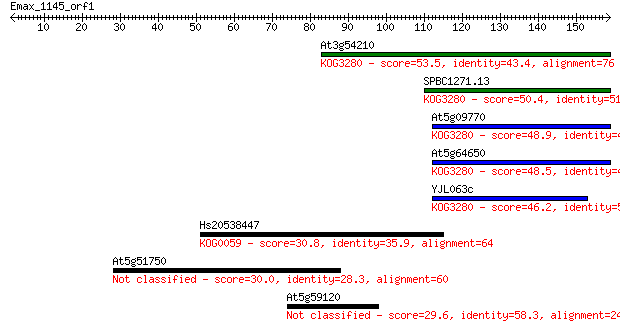
At3g54210 53.5 1e-07
SPBC1271.13 50.4 2e-06
At5g09770 48.9 4e-06
At5g64650 48.5 5e-06
YJL063c 46.2 2e-05
Hs20538447 30.8 1.1
At5g51750 30.0 2.0
At5g59120 29.6 2.8
> At3g54210
Length=211
Score = 53.5 bits (127), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 33/76 (43%), Positives = 45/76 (59%), Gaps = 4/76 (5%)
Query 83 THQSRFKKPLICGGPLTVVMRAHVRRSHSRLGRPRSHRKALLRNLATQLIRHGSIITTAA 142
+ ++ FK ++ GG MR H RR +L RP RKALLR L TQL++HG I TT A
Sbjct 79 SFENGFK--IVDGGGRIYAMR-HGRRV-PKLNRPPDQRKALLRGLTTQLLKHGRIKTTRA 134
Query 143 KGKELRRVIDSKIILA 158
+ +R+ +D I LA
Sbjct 135 RASAMRKFVDKMITLA 150
> SPBC1271.13
Length=207
Score = 50.4 bits (119), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 25/49 (51%), Positives = 34/49 (69%), Gaps = 0/49 (0%)
Query 110 HSRLGRPRSHRKALLRNLATQLIRHGSIITTAAKGKELRRVIDSKIILA 158
+ +LGR +HR+ALLRNL T L++H SI TT AK KE +R + I +A
Sbjct 7 YRKLGRTSAHRQALLRNLVTSLVKHESIQTTWAKAKEAQRFAEKLITMA 55
> At5g09770
Length=156
Score = 48.9 bits (115), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 22/47 (46%), Positives = 32/47 (68%), Gaps = 0/47 (0%)
Query 112 RLGRPRSHRKALLRNLATQLIRHGSIITTAAKGKELRRVIDSKIILA 158
+LGRP HR ++LR + +QL++H I TT K KE+RR+ D+ I L
Sbjct 6 KLGRPAGHRMSMLRTMVSQLVKHERIETTVTKAKEVRRLADNMIQLG 52
> At5g64650
Length=156
Score = 48.5 bits (114), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 22/47 (46%), Positives = 32/47 (68%), Gaps = 0/47 (0%)
Query 112 RLGRPRSHRKALLRNLATQLIRHGSIITTAAKGKELRRVIDSKIILA 158
+LGRP HR ++LR + +QL++H I TT K KE+RR+ D+ I L
Sbjct 6 KLGRPAGHRMSMLRTMVSQLVQHERIETTVTKAKEVRRLADNMIQLG 52
> YJL063c
Length=238
Score = 46.2 bits (108), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 22/41 (53%), Positives = 30/41 (73%), Gaps = 0/41 (0%)
Query 112 RLGRPRSHRKALLRNLATQLIRHGSIITTAAKGKELRRVID 152
+L R ++HR ALL+NLA QL +H SI++T AK KE RV +
Sbjct 8 KLSRDKAHRDALLKNLACQLFQHESIVSTHAKCKEASRVAE 48
> Hs20538447
Length=4273
Score = 30.8 bits (68), Expect = 1.1, Method: Composition-based stats.
Identities = 23/68 (33%), Positives = 36/68 (52%), Gaps = 10/68 (14%)
Query 51 CLKMDKQTTLRNGRPEVITNSYCS----PWLCTVSCTHQSRFKKPLICGGPLTVVMRAHV 106
CL+ ++T + G E NS+CS P+L T+S QS +K PL+ G + +H
Sbjct 3625 CLQRGRETMVGLGLSEPEVNSHCSQQKFPFLMTLSA--QSCWKPPLLPAGSVL----SHQ 3678
Query 107 RRSHSRLG 114
RR + +G
Sbjct 3679 RRFLNNVG 3686
> At5g51750
Length=780
Score = 30.0 bits (66), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 17/60 (28%), Positives = 28/60 (46%), Gaps = 7/60 (11%)
Query 28 ASRTRSASSINIFSRSPQAMHVSCLKMDKQTTLRNGRPEVITNSYCSPWLCTVSCTHQSR 87
++ +R + SI F + VSC + NG P+ I+ + SPW+ TV + R
Sbjct 302 STYSRDSLSIATFGAMEMGVFVSC-------SAGNGGPDPISLTNVSPWITTVGASTMDR 354
> At5g59120
Length=732
Score = 29.6 bits (65), Expect = 2.8, Method: Composition-based stats.
Identities = 14/25 (56%), Positives = 17/25 (68%), Gaps = 1/25 (4%)
Query 74 SPWLCTVSCTHQSRFK-KPLICGGP 97
S LC +SC +SR K K L+CGGP
Sbjct 370 SAGLCELSCVDKSRVKGKILVCGGP 394
Lambda K H
0.323 0.130 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2100092188
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40