bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_1146_orf1
Length=209
Score E
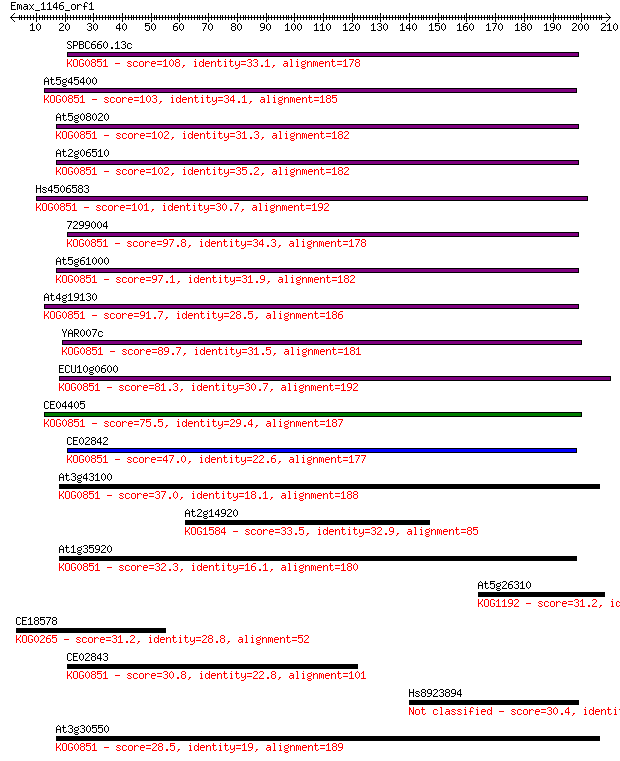
Sequences producing significant alignments: (Bits) Value
SPBC660.13c 108 6e-24
At5g45400 103 2e-22
At5g08020 102 6e-22
At2g06510 102 8e-22
Hs4506583 101 1e-21
7299004 97.8 1e-20
At5g61000 97.1 2e-20
At4g19130 91.7 1e-18
YAR007c 89.7 4e-18
ECU10g0600 81.3 1e-15
CE04405 75.5 8e-14
CE02842 47.0 3e-05
At3g43100 37.0 0.029
At2g14920 33.5 0.30
At1g35920 32.3 0.79
At5g26310 31.2 1.5
CE18578 31.2 1.7
CE02843 30.8 1.8
Hs8923894 30.4 2.4
At3g30550 28.5 9.4
> SPBC660.13c
Length=609
Score = 108 bits (271), Expect = 6e-24, Method: Compositional matrix adjust.
Identities = 59/182 (32%), Positives = 107/182 (58%), Gaps = 9/182 (4%)
Query 21 ISSLNPYVHRWQIKARVVDKSPLQ---TTKNNSKFFHVDVTDSAGDLIRAKFWGDAAEKW 77
I L+PY ++W I+ARV +KS ++ + K F V++ D +G+ IRA + D + +
Sbjct 182 IEGLSPYQNKWTIRARVTNKSEVKHWHNQRGEGKLFSVNLLDESGE-IRATGFNDQVDAF 240
Query 78 HGALEKGKVYCFSRGRVNVANKRFSTLSNNYELSFSAEADIKPAEDDGSIH-AKRSLCSM 136
+ L++G VY SR RVN+A K+++ + N YEL F + +I+ AED ++ AK S S
Sbjct 241 YDILQEGSVYYISRCRVNIAKKQYTNVQNEYELMFERDTEIRKAEDQTAVPVAKFSFVS- 299
Query 137 PLRGIFNSTRECPFPTDVLAVVVEVQPISSVTARSSGEELKRRVLKVRDKSQMEMEISLW 196
L+ + + ++ DV+ V+ V P+ +T+R++ +R + + D++ EM ++LW
Sbjct 300 -LQEVGDVAKDAVI--DVIGVLQNVGPVQQITSRATSRGFDKRDITIVDQTGYEMRVTLW 356
Query 197 GE 198
G+
Sbjct 357 GK 358
> At5g45400
Length=853
Score = 103 bits (258), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 63/189 (33%), Positives = 99/189 (52%), Gaps = 11/189 (5%)
Query 13 PPCERFLSISSLNPYVHRWQIKARVVDKSPLQTTKN---NSKFFHVDVTDSAGDLIRAKF 69
PP R I++LNPY RW IK RV K+ L+ N K F D+ D+ G IR
Sbjct 296 PP--RINPIAALNPYQGRWTIKVRVTSKADLRRFNNPRGEGKLFSFDLLDADGGEIRVTC 353
Query 70 WGDAAEKWHGALEKGKVYCFSRGRVNVANKRFSTLSNNYELSFSAEADIKPAEDDGSIHA 129
+ DA +++ + G VY SRG + A K F+ L N+YE+ + + I+P EDDG+I
Sbjct 354 FNDAVDQFFDKIVVGNVYLISRGNLKPAQKNFNHLPNDYEIHLDSASTIQPCEDDGTIPR 413
Query 130 KRSLCSMPLRGIFN-STRECPFPTDVLAVVVEVQPISSVTARSSGEELKRRVLKVRDKSQ 188
R I + E TDV+ +V + P ++ R + E+++R L+++D S
Sbjct 414 YH----FHFRNIGDIENMENNSTTDVIGIVSSISPTVAIM-RKNLTEVQKRSLQLKDMSG 468
Query 189 MEMEISLWG 197
+E+++WG
Sbjct 469 RSVEVTMWG 477
> At5g08020
Length=604
Score = 102 bits (254), Expect = 6e-22, Method: Compositional matrix adjust.
Identities = 57/187 (30%), Positives = 103/187 (55%), Gaps = 8/187 (4%)
Query 17 RFLSISSLNPYVHRWQIKARVVDKSPLQTTKN---NSKFFHVDVTDSAGDLIRAKFWGDA 73
R + SLNPY W IK RV +K ++T KN F+V++TD G I+A + A
Sbjct 156 RVHPLVSLNPYQGSWTIKVRVTNKGVMRTYKNARGEGCVFNVELTDEEGTQIQATMFNAA 215
Query 74 AEKWHGALEKGKVYCFSRGRVNVANKRFSTLSNNYELSFSAEADIKPAEDDGSIHAKRSL 133
A K++ E GKVY SRG + +ANK+F T+ N+YE++ + ++++ A ++ +
Sbjct 216 ARKFYDRFEMGKVYYISRGSLKLANKQFKTVQNDYEMTLNENSEVEEASNEEMFTPETKF 275
Query 134 CSMPLR--GIFNSTRECPFPTDVLAVVVEVQPISSVTARSSGEELKRRVLKVRDKSQMEM 191
+P+ G + + ++ DV+ VV V P S+ ++ E + +R + + D+++ +
Sbjct 276 NFVPIDELGTYVNQKDL---IDVIGVVQSVSPTMSIRRKNDNEMIPKRDITLADETKKTV 332
Query 192 EISLWGE 198
+SLW +
Sbjct 333 VVSLWND 339
> At2g06510
Length=640
Score = 102 bits (253), Expect = 8e-22, Method: Compositional matrix adjust.
Identities = 64/186 (34%), Positives = 97/186 (52%), Gaps = 8/186 (4%)
Query 17 RFLSISSLNPYVHRWQIKARVVDKSPLQ---TTKNNSKFFHVDVTDSAGDLIRAKFWGDA 73
R + I++LNPY RW IKARV K ++ K + K F D+ D G IR +
Sbjct 196 RVIPIAALNPYQGRWAIKARVTAKGDIRRYNNAKGDGKVFSFDLLDYDGGEIRVTCFNAL 255
Query 74 AEKWHGALEKGKVYCFSRGRVNVANKRFSTLSNNYELSFSAEADIKPAED-DGSIHAKRS 132
++++ E GKVY S+G + A K F+ L N +E+ + + ++ D DGSI K+
Sbjct 256 VDRFYDVTEVGKVYLISKGSLKPAQKNFNHLKNEWEIFLESTSTVELCPDEDGSI-PKQQ 314
Query 133 LCSMPLRGIFNSTRECPFPTDVLAVVVEVQPISSVTARSSGEELKRRVLKVRDKSQMEME 192
P+ I N+ DV+ VV V P S R +G E RR+L ++D+S +E
Sbjct 315 FSFRPISDIENAENNTIL--DVIGVVTSVNP-SVPILRKNGMETHRRILNLKDESGKAVE 371
Query 193 ISLWGE 198
++LWGE
Sbjct 372 VTLWGE 377
> Hs4506583
Length=616
Score = 101 bits (251), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 59/196 (30%), Positives = 105/196 (53%), Gaps = 10/196 (5%)
Query 10 SGGPPCERFLSISSLNPYVHRWQIKARVVDKSPLQT---TKNNSKFFHVDVTDSAGDLIR 66
SGG + + I+SL PY +W I ARV +KS ++T ++ K F +++ D +G+ IR
Sbjct 177 SGGTQ-SKVVPIASLTPYQSKWTICARVTNKSQIRTWSNSRGEGKLFSLELVDESGE-IR 234
Query 67 AKFWGDAAEKWHGALEKGKVYCFSRGRVNVANKRFSTLSNNYELSFSAEADIKPAEDDGS 126
A + + +K+ +E KVY FS+G + +ANK+F+ + N+YE++F+ E + P EDD
Sbjct 235 ATAFNEQVDKFFPLIEVNKVYYFSKGTLKIANKQFTAVKNDYEMTFNNETSVMPCEDDHH 294
Query 127 IHAKRSLCSMPLRGIFN-STRECPFPTDVLAVVVEVQPISSVTARSSGEELKRRVLKVRD 185
+ GI + + D++ + + + +T RS+ E+ +R + + D
Sbjct 295 LPT----VQFDFTGIDDLENKSKDSLVDIIGICKSYEDATKITVRSNNREVAKRNIYLMD 350
Query 186 KSQMEMEISLWGEQVD 201
S + +LWGE D
Sbjct 351 TSGKVVTATLWGEDAD 366
> 7299004
Length=603
Score = 97.8 bits (242), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 61/184 (33%), Positives = 105/184 (57%), Gaps = 11/184 (5%)
Query 21 ISSLNPYVHRWQIKARVVDKSPLQTTKN---NSKFFHVDVTDSAGDLIRAKFWGDAAEKW 77
ISSL+PY ++W IKARV KS ++T N K F +D+ D +G+ IRA + + +K+
Sbjct 169 ISSLSPYQNKWVIKARVTSKSGIRTWSNARGEGKLFSMDLMDESGE-IRATAFKEQCDKF 227
Query 78 HGALEKGKVYCFSRGRVNVANKRFSTLSNNYELSFSAEADIKPAE---DDGSIHAKRSLC 134
+ ++ VY S+ ++ ANK++S+L+N YE++FS E ++ E DD K +L
Sbjct 228 YDLIQVDSVYYISKCQLKPANKQYSSLNNAYEMTFSGETVVQLCEDTDDDPIPEIKYNL- 286
Query 135 SMPLRGIFNSTRECPFPTDVLAVVVEVQPISSVTARSSGEELKRRVLKVRDKSQMEMEIS 194
+P+ + S E D + + EV + S AR++ +E K+R + + D S + ++
Sbjct 287 -VPISDV--SGMENKAAVDTIGICKEVGELQSFVARTTNKEFKKRDITLVDMSNSAISLT 343
Query 195 LWGE 198
LWG+
Sbjct 344 LWGD 347
> At5g61000
Length=629
Score = 97.1 bits (240), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 58/187 (31%), Positives = 102/187 (54%), Gaps = 8/187 (4%)
Query 17 RFLSISSLNPYVHRWQIKARVVDKSPLQTTKN---NSKFFHVDVTDSAGDLIRAKFWGDA 73
R + SLNPY W IK RV +K ++ KN F+V++TD G I+A + DA
Sbjct 180 RVHPLVSLNPYQGNWTIKVRVTNKGVMRNYKNARGEGCVFNVELTDEEGTQIQATMFNDA 239
Query 74 AEKWHGALEKGKVYCFSRGRVNVANKRFSTLSNNYELSFSAEADIKPAEDDGSIHAKRSL 133
A K+ + GKVY SRG + +ANK+F T+ N+YE++ + ++++ A + +
Sbjct 240 ARKFFDRFQLGKVYYISRGSLKLANKQFKTVQNDYEMTLNENSEVEEASSEEMFIPETKF 299
Query 134 CSMPLR--GIFNSTRECPFPTDVLAVVVEVQPISSVTARSSGEELKRRVLKVRDKSQMEM 191
+P+ G++ + +E D++ VV V P S+ R+ E + +R + + D+S+ +
Sbjct 300 NFVPIEELGLYVNQKEL---IDLIGVVQSVSPTMSIRRRTDNEMIPKRDITLADESRKTV 356
Query 192 EISLWGE 198
+SLW +
Sbjct 357 VVSLWND 363
> At4g19130
Length=785
Score = 91.7 bits (226), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 53/189 (28%), Positives = 98/189 (51%), Gaps = 8/189 (4%)
Query 13 PPCERFLSISSLNPYVHRWQIKARVVDKSPLQTTKN---NSKFFHVDVTDSAGDLIRAKF 69
PP + + +++L+PY RW IKARV +K+ L+ N K F+ D+ D+ G IR
Sbjct 226 PP--KIIPVNALSPYSGRWTIKARVTNKAALKQYSNPRGEGKVFNFDLLDADGGEIRVTC 283
Query 70 WGDAAEKWHGALEKGKVYCFSRGRVNVANKRFSTLSNNYELSFSAEADIKPAEDDGSIHA 129
+ A++++ + G +Y SRG + A K F+ L N+YE+ + IK ++ +
Sbjct 284 FNAVADQFYDQIVVGNLYLISRGSLRPAQKNFNHLRNDYEIMLDNASTIKQCYEEDAAIP 343
Query 130 KRSLCSMPLRGIFNSTRECPFPTDVLAVVVEVQPISSVTARSSGEELKRRVLKVRDKSQM 189
+ + I + C DV+ +V + P ++T R +G +R L+++D S
Sbjct 344 RHQFHFRTIGDIESMENNC--IVDVIGIVSSISPTVTIT-RKNGTATPKRSLQLKDMSGR 400
Query 190 EMEISLWGE 198
+E+++WG+
Sbjct 401 SVEVTMWGD 409
> YAR007c
Length=621
Score = 89.7 bits (221), Expect = 4e-18, Method: Compositional matrix adjust.
Identities = 57/184 (30%), Positives = 101/184 (54%), Gaps = 8/184 (4%)
Query 19 LSISSLNPYVHRWQIKARVVDKSPLQTTKN---NSKFFHVDVTDSAGDLIRAKFWGDAAE 75
+I L+PY + W IKARV K ++T N + K F+V+ D++G+ IRA + D A
Sbjct 185 FAIEQLSPYQNVWTIKARVSYKGEIKTWHNQRGDGKLFNVNFLDTSGE-IRATAFNDFAT 243
Query 76 KWHGALEKGKVYCFSRGRVNVANKRFSTLSNNYELSFSAEADIKPAEDDGSIHAKRSLCS 135
K++ L++GKVY S+ ++ A +F+ L++ YEL+ + I+ D+ ++ K
Sbjct 244 KFNEILQEGKVYYVSKAKLQPAKPQFTNLTHPYELNLDRDTVIEECFDESNV-PKTHFNF 302
Query 136 MPLRGIFNSTRECPFPTDVLAVVVEVQPISSVTARSSGEELKRRVLKVRDKSQMEMEISL 195
+ L I N +E DVL ++ + P +T+R +G++ RR + + D S + + L
Sbjct 303 IKLDAIQN--QEVNSNVDVLGIIQTINPHFELTSR-AGKKFDRRDITIVDDSGFSISVGL 359
Query 196 WGEQ 199
W +Q
Sbjct 360 WNQQ 363
> ECU10g0600
Length=623
Score = 81.3 bits (199), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 59/196 (30%), Positives = 99/196 (50%), Gaps = 13/196 (6%)
Query 18 FLSISSLNPYVHRWQIKARVVDKSPLQ---TTKNNSKFFHVDVTDSAGDLIRAKFWGDAA 74
F +I+ LNP+ ++W IK RVV KS ++ K K F+ +V+D ++ + D
Sbjct 214 FTAINMLNPFHNKWAIKGRVVMKSDIRRFTNQKGEGKVFNFEVSDGTAQ-VKIICFSDCV 272
Query 75 EKWHGALEKGKVYCFSRGRVNVANKRFSTLSNNYELSFSAEADIKPAEDDGSIHAKRSLC 134
+ + +E GKVY ++G V +ANK++ST +YE+ +++ A DDGS K
Sbjct 273 DIFFPIVEVGKVYTIAKGTVKMANKQYSTNPFDYEIILDKSSEVHRAADDGS--PKYFFN 330
Query 135 SMPLRGIFNSTRECPFPTDVLAVVVEVQPISSVTARSSGEE-LKRRVLKVRDKSQMEMEI 193
+ + + C D + VV EV S+V RS+ E LKR + V D + +
Sbjct 331 FVKISDLTLGNAYC----DTIGVVKEVYAPSTVMVRSTQSELLKRDAVLVDDGGSVRL-- 384
Query 194 SLWGEQVDCITETTAI 209
+LWG + + E+ +
Sbjct 385 TLWGPKAELEIESGMV 400
> CE04405
Length=655
Score = 75.5 bits (184), Expect = 8e-14, Method: Compositional matrix adjust.
Identities = 55/198 (27%), Positives = 99/198 (50%), Gaps = 17/198 (8%)
Query 13 PPCER--------FLSISSLNPYVHRWQIKARVVDKSPLQT-TKNNSKFFHVDVTDSAGD 63
PP R + I+ + PYV ++I V K ++T N+K F+ ++TDS GD
Sbjct 210 PPARRTASNTERGVMPIAMVTPYVSNFKIHGMVSRKEEIRTFPAKNTKVFNFEITDSNGD 269
Query 64 LIRAKFWGDAAEKWHGALEKGKVYCFSRGRVNVANKRFSTLSNNYELSFSAEADIKPAED 123
IR + + AE + + + Y S G V ANK+F+ ++YE++ +++ I E
Sbjct 270 TIRCTAFNEVAESLYTTITENLSYYLSGGSVKQANKKFNNTGHDYEITLRSDSII---EA 326
Query 124 DGSIHA--KRSLCSMPLRGIFNSTRECPFPTDVLAVVVEVQPISSVTARSSGEELKRRVL 181
G + A K L + L I + DVL VV ++ P ++ +G+ L +R +
Sbjct 327 GGELLAAPKLILKRVKLGEIAGYAGQL---IDVLVVVEKMDPEATEFTSKAGKSLIKREM 383
Query 182 KVRDKSQMEMEISLWGEQ 199
++ D+S + ++LWG++
Sbjct 384 ELIDESGALVRLTLWGDE 401
> CE02842
Length=391
Score = 47.0 bits (110), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 40/177 (22%), Positives = 77/177 (43%), Gaps = 5/177 (2%)
Query 21 ISSLNPYVHRWQIKARVVDKSPLQTTKNNSKFFHVDVTDSAGDLIRAKFWGDAAEKWHGA 80
I++LN V ++I RV + + + K F ++TD G IR +G+ + +G+
Sbjct 190 IANLNDSVDNFKIHGRV---TLMDDKRRPEKVFSFEITDVNGYTIRCVAFGELGVRLYGS 246
Query 81 LEKGKVYCFSRGRVNVANKRFSTLSNNYELSFSAEADIKPAEDDGSIHAKRSLCSMPLRG 140
+ K + Y + G+V + ++ + +E+ I+PA ++ K +L + L
Sbjct 247 IAKDQSYYLTGGKVKNGHTLYNQTGHAFEIILDEFPTIEPAPAVLTV-PKLNLNRVMLCN 305
Query 141 IFNSTRECPFPTDVLAVVVEVQPISSVTARSSGEELKRRVLKVRDKSQMEMEISLWG 197
+ N P DV+ V E+ + R + V D S + ++LWG
Sbjct 306 VQNEQYYRK-PIDVIVAVEEINDFLDDYHPIENKPPVLRNMVVIDDSNFRIRLTLWG 361
> At3g43100
Length=269
Score = 37.0 bits (84), Expect = 0.029, Method: Compositional matrix adjust.
Identities = 34/188 (18%), Positives = 77/188 (40%), Gaps = 6/188 (3%)
Query 18 FLSISSLNPYVHRWQIKARVVDKSPLQTTKNNSKFFHVDVTDSAGDLIRAKFWGDAAEKW 77
F + SL PY + W+I+ +++ + K + + D AGD + A + +K+
Sbjct 9 FTPLKSLKPYKNAWRIQVKLLHVWRQYSVKAGESIEMI-LVDEAGDKMYAAVRREQIKKF 67
Query 78 HGALEKGKVYCFSRGRVNVANKRFSTLSNNYELSFSAEADIKPAEDDGSIHAKRSLCSMP 137
L +G + +N + ++ Y++ F + + P + S+ L
Sbjct 68 ERCLTEGVWKIITTITLNPTSGKYRISDLKYKIGFVFKTTVSPCD---SVSDALFLSLAK 124
Query 138 LRGIFNSTRECPFPTDVLAVVVEVQPISSVTARSSGEELKRRVLKVRDKSQMEMEISLWG 197
I + + DV+ VV+ I + A + + K+ ++D+ + +LWG
Sbjct 125 FDVILSGSANSNILHDVMGQVVDRSEIQDLNA--NNKPTKKIDFHLKDQHDTRLACTLWG 182
Query 198 EQVDCITE 205
+ + + +
Sbjct 183 KYAEIVDQ 190
> At2g14920
Length=333
Score = 33.5 bits (75), Expect = 0.30, Method: Compositional matrix adjust.
Identities = 28/89 (31%), Positives = 37/89 (41%), Gaps = 9/89 (10%)
Query 62 GDLIRAKFWGDAAEKWHGALEKGKVYCFSRGRVNVANKRFST--LSNNYELSFSAEADIK 119
G + FW A W G+LE K + F R A R L+ + F+ E
Sbjct 204 GVTLHGPFWDHALSYWRGSLEDPKHFLFMRYEDLKAEPRTQVKRLAEFLDCPFTKE---- 259
Query 120 PAEDDGSIHAKRSLCSMP-LRGI-FNSTR 146
ED GS+ LCS+ LR + N TR
Sbjct 260 -EEDSGSVDKILELCSLSNLRSVEINKTR 287
> At1g35920
Length=567
Score = 32.3 bits (72), Expect = 0.79, Method: Compositional matrix adjust.
Identities = 29/180 (16%), Positives = 74/180 (41%), Gaps = 6/180 (3%)
Query 18 FLSISSLNPYVHRWQIKARVVDKSPLQTTKNNSKFFHVDVTDSAGDLIRAKFWGDAAEKW 77
F + + PY + W+++ +++ S Q T N + + ++D G + A + K+
Sbjct 5 FAYLKDVRPYKNAWRVQVKIL-HSWKQYTSNTRETIELVISDEHGKKMHATVKKELVSKF 63
Query 78 HGALEKGKVYCFSRGRVNVANKRFSTLSNNYELSFSAEADIKPAEDDGSIHAKRSLCSMP 137
L G+ + A+ +F ++ Y+++F ++ S+ L
Sbjct 64 VHKLIVGEWVFIEIFGLTYASGQFRPTNHLYKMAFQVRTEVMGC---ASVSDSNFLTLAS 120
Query 138 LRGIFNSTRECPFPTDVLAVVVEVQPISSVTARSSGEELKRRVLKVRDKSQMEMEISLWG 197
I + D + ++ V + + +++ + + ++RD+ M+++LWG
Sbjct 121 FSKIQSGELNPHMLVDAIGQIITVGELEEL--KANNKRTTKIDFEIRDQMDERMQVTLWG 178
> At5g26310
Length=481
Score = 31.2 bits (69), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 14/44 (31%), Positives = 24/44 (54%), Gaps = 0/44 (0%)
Query 164 ISSVTARSSGEELKRRVLKVRDKSQMEMEISLWGEQVDCITETT 207
+ V A GEE++R+V K+RD ++M + I G + + T
Sbjct 421 VRKVMAEDEGEEMRRKVKKLRDTAEMSLSIHGGGSAHESLCRVT 464
> CE18578
Length=331
Score = 31.2 bits (69), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 15/53 (28%), Positives = 26/53 (49%), Gaps = 1/53 (1%)
Query 3 FATKMLDSGGPPCERFLSISSLNPYVHRWQ-IKARVVDKSPLQTTKNNSKFFH 54
F +L PCERF++ S + +++ W+ + ++V K P N FH
Sbjct 256 FEKNLLKCSWSPCERFITAGSSDRFLYVWETLSKKIVYKLPGHMGSVNCTDFH 308
> CE02843
Length=299
Score = 30.8 bits (68), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 23/103 (22%), Positives = 48/103 (46%), Gaps = 5/103 (4%)
Query 21 ISSLNPYVHRWQIKARV--VDKSPLQTTKNNSKFFHVDVTDSAGDLIRAKFWGDAAEKWH 78
I +L V+ ++I RV +D P + K F+ ++TD GD I A+ ++
Sbjct 117 IENLKESVNYFKIHGRVTLMDTRP---PHHYQKRFNFEITDFNGDTIACSASSPEADVFY 173
Query 79 GALEKGKVYCFSRGRVNVANKRFSTLSNNYELSFSAEADIKPA 121
L K + Y + G + ++ ++ +Y++ + ++PA
Sbjct 174 QNLVKEQSYYLAGGHIKKGHEIYNQTGIDYQIDLNNFTIMEPA 216
> Hs8923894
Length=852
Score = 30.4 bits (67), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 20/60 (33%), Positives = 31/60 (51%), Gaps = 5/60 (8%)
Query 140 GIFNSTRECPFPTDVLAVVVEVQPISSVT-ARSSGEELKRRVLKVRDKSQMEMEISLWGE 198
G+F S CPFP D +VV+V +S+ AR + R+ L++R + E+ W E
Sbjct 148 GLFTSQPLCPFPVDKPNIVVDVIFTNSLPEARRN----SRQPLEIRTSKRTELAQGQWVE 203
> At3g30550
Length=509
Score = 28.5 bits (62), Expect = 9.4, Method: Compositional matrix adjust.
Identities = 36/196 (18%), Positives = 76/196 (38%), Gaps = 28/196 (14%)
Query 17 RFLSISSLNPYVHRWQIKARVVDKSPLQTTKNNSKFFHV--DVTDSAGDLIRAKFWGDAA 74
+F + SL PY + W+I+ K HV + AG+ I +A
Sbjct 8 QFTPLKSLKPYKNAWRIQV---------------KLLHVWRQYSVKAGESIEIILVDEAG 52
Query 75 EKWHGALEKGKVYCFSRGRVNVANKRFSTLSNN-----YELSFSAEADIKPAEDDGSIHA 129
+K + ++ + ++ F R K +T++ N Y F + + P ++ +
Sbjct 53 DKMYASVRREQIKKFERCLTEGVWKIITTITLNPTSGQYHW-FCFQTTVSPCDN---VSD 108
Query 130 KRSLCSMPLRGIFNSTRECPFPTDVLAVVVEVQPISSVTARSSGEELKRRVLKVRDKSQM 189
L I + + +++ VV I ++ A S + K+ +RD+
Sbjct 109 ALFLSLAKFDVILSRSANSNILHNIIGQVVNRSEIQNLNA--SNKPTKKIDFHLRDQHDT 166
Query 190 EMEISLWGEQVDCITE 205
+ +LWG+ + + +
Sbjct 167 RLACTLWGKYAEIVDQ 182
Lambda K H
0.318 0.132 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3754464630
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40