bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_1169_orf1
Length=123
Score E
Sequences producing significant alignments: (Bits) Value
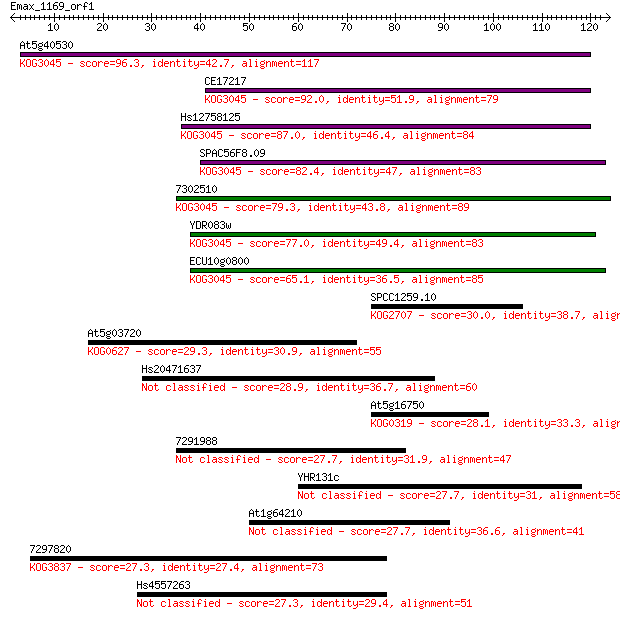
At5g40530 96.3 1e-20
CE17217 92.0 2e-19
Hs12758125 87.0 8e-18
SPAC56F8.09 82.4 2e-16
7302510 79.3 2e-15
YDR083w 77.0 8e-15
ECU10g0800 65.1 3e-11
SPCC1259.10 30.0 0.96
At5g03720 29.3 1.9
Hs20471637 28.9 2.7
At5g16750 28.1 4.4
7291988 27.7 5.4
YHR131c 27.7 6.0
At1g64210 27.7 6.1
7297820 27.3 6.7
Hs4557263 27.3 6.8
> At5g40530
Length=285
Score = 96.3 bits (238), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 50/128 (39%), Positives = 71/128 (55%), Gaps = 11/128 (8%)
Query 3 PSADEKRETMANLQQQKERQLLLVKKFSGRVAA-------AGVID----RLQGGRFRSLN 51
PS +E ET Q +K+ Q + G +A + +D RL GG+FR LN
Sbjct 22 PSKEEPIETTPKNQNEKKNQRDTKNQQHGGSSAPSKRPKPSNFLDALRERLSGGQFRMLN 81
Query 52 EYLYTKKGSEAFVKYQKEPQLFDIYHTGYRAQVRRWPLNPLDCVATWLSKKPKEWVVGDF 111
E LYT G EA ++++PQ+FD+YHTGY+ Q+ WP P++ + WL VV DF
Sbjct 82 EKLYTCSGKEALDYFKEDPQMFDMYHTGYQQQMSNWPELPVNSIINWLLSNSSSLVVADF 141
Query 112 GCGEAALA 119
GCG+A +A
Sbjct 142 GCGDARIA 149
> CE17217
Length=343
Score = 92.0 bits (227), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 41/79 (51%), Positives = 53/79 (67%), Gaps = 0/79 (0%)
Query 41 RLQGGRFRSLNEYLYTKKGSEAFVKYQKEPQLFDIYHTGYRAQVRRWPLNPLDCVATWLS 100
RL GRFR LNE LYT GSEAF ++++P FD+YH G+ QV++WP +PL + WL
Sbjct 133 RLDAGRFRFLNEKLYTCTGSEAFDFFKEDPTAFDLYHKGFADQVKKWPNHPLREIIRWLQ 192
Query 101 KKPKEWVVGDFGCGEAALA 119
KP + V D GCGEA +A
Sbjct 193 SKPDQQSVFDLGCGEAKIA 211
> Hs12758125
Length=456
Score = 87.0 bits (214), Expect = 8e-18, Method: Compositional matrix adjust.
Identities = 39/84 (46%), Positives = 56/84 (66%), Gaps = 0/84 (0%)
Query 36 AGVIDRLQGGRFRSLNEYLYTKKGSEAFVKYQKEPQLFDIYHTGYRAQVRRWPLNPLDCV 95
A + RL G RFR LNE LY+ S A +Q++P+ F +YH G+++QV++WPL P+D +
Sbjct 240 ARMAQRLDGARFRYLNEQLYSGPSSAAQRLFQEDPEAFLLYHRGFQSQVKKWPLQPVDRI 299
Query 96 ATWLSKKPKEWVVGDFGCGEAALA 119
A L ++P VV DFGCG+ LA
Sbjct 300 ARDLRQRPASLVVADFGCGDCRLA 323
> SPAC56F8.09
Length=318
Score = 82.4 bits (202), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 39/91 (42%), Positives = 51/91 (56%), Gaps = 8/91 (8%)
Query 40 DRLQGGRFRSLNEYLYTKKGSEAFVKYQKEPQLFDIYHTGYRAQVRRWPLNPLDCVATWL 99
D+L G FR +NE LYT + +A +++ P LFDIYH G+R QV WP NP+D L
Sbjct 95 DKLDGANFRWINEQLYTTESDKAVQMFKENPDLFDIYHAGFRYQVEGWPENPVDIFIQHL 154
Query 100 --------SKKPKEWVVGDFGCGEAALALRF 122
+KK V+ D GCGEA +A F
Sbjct 155 KIRFEHSNAKKKNNIVIADLGCGEAKIASTF 185
> 7302510
Length=358
Score = 79.3 bits (194), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 39/89 (43%), Positives = 55/89 (61%), Gaps = 0/89 (0%)
Query 35 AAGVIDRLQGGRFRSLNEYLYTKKGSEAFVKYQKEPQLFDIYHTGYRAQVRRWPLNPLDC 94
A+ + L GGRFR +NE LY+ +A ++K+ F+ YH GYR QV +WP+NPL+
Sbjct 143 ASKLQSELLGGRFRYINEQLYSTTSRKAEALFRKDSSAFEAYHAGYRQQVEKWPINPLNR 202
Query 95 VATWLSKKPKEWVVGDFGCGEAALALRFP 123
+ + K PK ++GDFGCGE LA P
Sbjct 203 IIKTIKKIPKTAIIGDFGCGEGKLAQSVP 231
> YDR083w
Length=402
Score = 77.0 bits (188), Expect = 8e-15, Method: Composition-based stats.
Identities = 41/99 (41%), Positives = 54/99 (54%), Gaps = 16/99 (16%)
Query 38 VIDRLQGGRFRSLNEYLYTKKGSEAFVKYQKEPQLFDIYHTGYRAQVRRWPLNPLDCVAT 97
++ +L G RFR +NE LYT EA +++PQLFD YH G+R+QV+ WP NP+D
Sbjct 136 MMAKLTGSRFRWINEQLYTISSDEALKLIKEQPQLFDEYHDGFRSQVQAWPENPVDVFVD 195
Query 98 WLS----------------KKPKEWVVGDFGCGEAALAL 120
+ K KE V+ D GCGEA LAL
Sbjct 196 QIRYRCMKPVNAPGGLPGLKDSKEIVIADMGCGEAQLAL 234
> ECU10g0800
Length=210
Score = 65.1 bits (157), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 31/85 (36%), Positives = 45/85 (52%), Gaps = 8/85 (9%)
Query 38 VIDRLQGGRFRSLNEYLYTKKGSEAFVKYQKEPQLFDIYHTGYRAQVRRWPLNPLDCVAT 97
+ RL+GG+FR LN+ +Y KG +YH Y QV +WP+NPLD +
Sbjct 7 ITKRLEGGKFRMLNDKMYHGKGLRK--------GDLRLYHELYNLQVMKWPVNPLDVIIE 58
Query 98 WLSKKPKEWVVGDFGCGEAALALRF 122
+ ++ V+ D GCGEA +A F
Sbjct 59 KIKRREGNGVIADIGCGEARIAREF 83
> SPCC1259.10
Length=412
Score = 30.0 bits (66), Expect = 0.96, Method: Composition-based stats.
Identities = 12/36 (33%), Positives = 21/36 (58%), Gaps = 5/36 (13%)
Query 75 IYHTGYRAQ-----VRRWPLNPLDCVATWLSKKPKE 105
++ GY + +R+WP+N + V WL+KK K+
Sbjct 376 MFKAGYTSSFDVEPIRKWPINQILTVEGWLTKKNKK 411
> At5g03720
Length=476
Score = 29.3 bits (64), Expect = 1.9, Method: Composition-based stats.
Identities = 17/57 (29%), Positives = 32/57 (56%), Gaps = 6/57 (10%)
Query 17 QQKERQLL--LVKKFSGRVAAAGVIDRLQGGRFRSLNEYLYTKKGSEAFVKYQKEPQ 71
+Q+++QLL L K F R G ++RL+ + + L +K + F+K+ ++PQ
Sbjct 275 EQRQKQLLSFLAKLFQNR----GFLERLKNFKGKEKGGALGLEKARKKFIKHHQQPQ 327
> Hs20471637
Length=176
Score = 28.9 bits (63), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 22/64 (34%), Positives = 32/64 (50%), Gaps = 9/64 (14%)
Query 28 KFSGRVAAAGVIDRLQGGRFRSLNEYLYTKKGSEAFV-KYQKEPQLFDIYH---TGYRAQ 83
F+ +G+ R QGGR L+ +L + S+ + K Q E Q+ D +H TG
Sbjct 51 HFAHDSFTSGLETRYQGGR-HGLHHFLSSTSRSQLLILKPQAEVQMQDFFHLQPTG---- 105
Query 84 VRRW 87
VRRW
Sbjct 106 VRRW 109
> At5g16750
Length=876
Score = 28.1 bits (61), Expect = 4.4, Method: Composition-based stats.
Identities = 8/24 (33%), Positives = 14/24 (58%), Gaps = 0/24 (0%)
Query 75 IYHTGYRAQVRRWPLNPLDCVATW 98
++ G+ Q+R W L L C+ +W
Sbjct 75 LFSAGHSRQIRVWDLETLKCIRSW 98
> 7291988
Length=288
Score = 27.7 bits (60), Expect = 5.4, Method: Compositional matrix adjust.
Identities = 15/47 (31%), Positives = 23/47 (48%), Gaps = 9/47 (19%)
Query 35 AAGVIDRLQGGRFRSLNEYLYTKKGSEAFVKYQKEPQLFDIYHTGYR 81
AA + DR G+ + E+L YQ P+L+D++H YR
Sbjct 2 AAALPDRFSAGKRQFWREFL---------ALYQGMPELWDVHHLNYR 39
> YHR131c
Length=840
Score = 27.7 bits (60), Expect = 6.0, Method: Composition-based stats.
Identities = 18/66 (27%), Positives = 27/66 (40%), Gaps = 8/66 (12%)
Query 60 SEAFVKYQKEPQLFDIYHTG--------YRAQVRRWPLNPLDCVATWLSKKPKEWVVGDF 111
S+ F++ P L Y TG R + + LD T LSK P E ++
Sbjct 338 SQRFIRSNSRPDLIQRYSTGSSTNNNTTIRERSNTFTAGLLDHYCTGLSKTPTEALISSA 397
Query 112 GCGEAA 117
GE++
Sbjct 398 ASGESS 403
> At1g64210
Length=587
Score = 27.7 bits (60), Expect = 6.1, Method: Composition-based stats.
Identities = 15/41 (36%), Positives = 19/41 (46%), Gaps = 1/41 (2%)
Query 50 LNEYLYTKKGSEAFVKYQKEPQLFDIYHTGYRAQVRRWPLN 90
L Y Y+K A Y LF+I H G R + R PL+
Sbjct 365 LKAYYYSKDDKLAVYSYYNHGSLFEILH-GNRGRYHRVPLD 404
> 7297820
Length=816
Score = 27.3 bits (59), Expect = 6.7, Method: Compositional matrix adjust.
Identities = 20/79 (25%), Positives = 36/79 (45%), Gaps = 6/79 (7%)
Query 5 ADEKRETMANLQQQKERQLLLVK------KFSGRVAAAGVIDRLQGGRFRSLNEYLYTKK 58
D+ E +++Q +QL++ K K G VA + L + L+ Y+K+
Sbjct 578 TDDICEIGVRMEEQLAKQLMMCKNTRDHHKAMGDVAGMNRFENLALTVQKDLDLVRYSKR 637
Query 59 GSEAFVKYQKEPQLFDIYH 77
+E K+ E + F+I H
Sbjct 638 KNEPLPKFHYEKRSFNIVH 656
> Hs4557263
Length=357
Score = 27.3 bits (59), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 15/51 (29%), Positives = 28/51 (54%), Gaps = 0/51 (0%)
Query 27 KKFSGRVAAAGVIDRLQGGRFRSLNEYLYTKKGSEAFVKYQKEPQLFDIYH 77
+K ++A G +D L GR+R ++ + +EA V+ K P+L +Y+
Sbjct 31 EKIHRQLAQLGGLDALDVGRWRVSDDTVMHLATAEALVEAGKAPKLTQLYY 81
Lambda K H
0.320 0.136 0.417
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1191192512
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40