bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_1190_orf4
Length=749
Score E
Sequences producing significant alignments: (Bits) Value
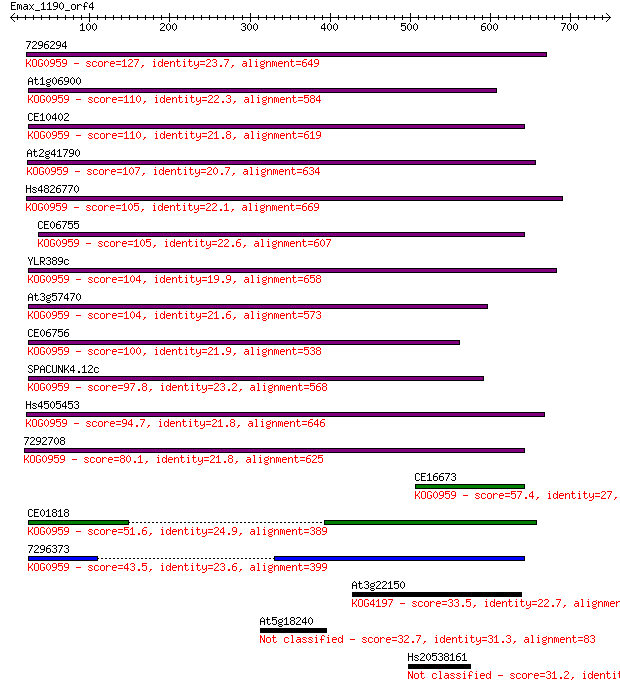
7296294 127 1e-28
At1g06900 110 8e-24
CE10402 110 9e-24
At2g41790 107 9e-23
Hs4826770 105 4e-22
CE06755 105 5e-22
YLR389c 104 6e-22
At3g57470 104 8e-22
CE06756 100 1e-20
SPACUNK4.12c 97.8 8e-20
Hs4505453 94.7 7e-19
7292708 80.1 2e-14
CE16673 57.4 1e-07
CE01818 51.6 6e-06
7296373 43.5 0.002
At3g22150 33.5 1.9
At5g18240 32.7 2.8
Hs20538161 31.2 9.2
> 7296294
Length=990
Score = 127 bits (318), Expect = 1e-28, Method: Compositional matrix adjust.
Identities = 154/671 (22%), Positives = 284/671 (42%), Gaps = 47/671 (7%)
Query 21 SLWVAFGLPATLTSYKKQPTSVLTYLLEYTGEGSLAKRLRLLGLADGISPAVDRNSISTL 80
SL ++F YK P + LT+L+ + G+GS+ LR LG + + A +N+ +
Sbjct 285 SLTISFTTDDLTQFYKSGPDNYLTHLIGHEGKGSILSELRRLGWCNDLM-AGHQNTQNGF 343
Query 81 --LGIKVDLTQKGAAHRGLVLQEIFSYINFLRDHGVGHELVSTMAQQSHIDFH---TTQP 135
I VDLTQ+G H +++ +F Y+ LR G + + + + F QP
Sbjct 344 GFFDIVVDLTQEGLEHVDDIVKIVFQYLEMLRKEGPKKWIFDECVKLNEMRFRFKEKEQP 403
Query 136 SSSIMDEAARLAHNLLTYEPYHVVAGDSLLIDADPRLTNQLLQEMSPSKAIIAFSDPDFT 195
+ + + + + V+ L + P L LL E+ PSK+ I F
Sbjct 404 ENLVTHAVSSMQ----IFPLEEVLIAPYLSNEWRPDLIKGLLDELVPSKSRIVIVSQSFE 459
Query 196 SKVDSFETDPYYGVQFRVLDLPQHHAVAMAVLTASPN-AFRMPPSLMHIPKASELKILPG 254
D E PYY ++ + + + + + N +P S IP ++ +P
Sbjct 460 PDCDLAE--PYYKTKYGITRVAKDTVQSWENCELNENLKLALPNSF--IPTNFDISDVPA 515
Query 255 LLGLTEPELISEQGGNAGTAVWWQGQGAFAIPRITVQLNGSILKDKADLLSRTQGSLALA 314
P +I + VW + F P+ + + S D L+ + +
Sbjct 516 D-APKHPTIILD---TPILRVWHKQDNQFNKPKACMTFDMSNPIAYLDPLNCNLNHMMVM 571
Query 315 AIAEHLQEETVDFQNCGVTHSLAFKGTGFHMGFEAYTEGQ---LSKLMEHVAKLLSDPSM 371
+ + L E D + + S+ K G +++ Q L KL++H L D S+
Sbjct 572 LLKDQLNEYLYDAELASLKLSVMGKSCGIDFTIRGFSDKQVVLLEKLLDH----LFDFSI 627
Query 372 VEPERFERIKQKQMKLVADPATSMAFEHALEAAAILTRNDAFSRKDLLNALQQTNYDD-- 429
E +RF+ +K++ ++ + + ++H++ A+L +A++ +LL+A++ YD
Sbjct 628 DE-KRFDILKEEYVRSLKNFKAEQPYQHSIYYLALLLTENAWANMELLDAMELVTYDRVL 686
Query 430 SFAKLNELKNVHVDAFVMGNIDRDQSLTM---VEEFLEQAGFTPIDHDDAVASLAMEQKQ 486
+FAK + +H + F+ GN+ + Q+ + V LE + + + + M +K+
Sbjct 687 NFAK-EFFQRLHTECFIFGNVTKQQATDIAGRVNTRLEATNASKL----PILARQMLKKR 741
Query 487 TIEATLANPIKGDKDH----ASLVQFQLGIPSIEDRVNLAV--LTQFLNRRIYDSLRTEA 540
+ + +K++ +S Q L + D N+ V ++Q L+ YD LRT+
Sbjct 742 EYKLLAGDSYLFEKENEFHKSSCAQLYLQCGAQTDHTNIMVNLVSQVLSEPCYDCLRTKE 801
Query 541 QLGYIAGAKESQAASTALLQCFVEGAKAHPDEVVKMIDEELSKAKEYLANMPEAEMARWK 600
QLGYI + + ++ V+ AK HP V I+ L + + +MP E R K
Sbjct 802 QLGYIVFSGVRKVNGANGIRIIVQSAK-HPSYVEDRIENFLQTYLQVIEDMPLDEFERHK 860
Query 601 EAAHAKLTKMEANFSEDFKKSAEEIFAHSNCFTKRDLEVKYLDNDFSRKQLLRTFAKL-- 658
EA K + + F + EI + F + + EV L S+ + F K
Sbjct 861 EALAVKKLEKPKTIFQQFSQFYGEIAMQTYHFEREEAEVAIL-RKISKADFVDYFKKFIA 919
Query 659 SDPSRRMVVKL 669
D R V+ +
Sbjct 920 KDGEERRVLSV 930
> At1g06900
Length=1023
Score = 110 bits (276), Expect = 8e-24, Method: Compositional matrix adjust.
Identities = 130/610 (21%), Positives = 250/610 (40%), Gaps = 49/610 (8%)
Query 24 VAFGLPATLTSYKKQPTSVLTYLLEYTGEGSLAKRLRLLGLADGISPAV-----DRNSIS 78
+ + LP ++Y K+P L +LL + G GSL L+ G A +S V +R+S++
Sbjct 335 LTWTLPPLRSAYVKKPEDYLAHLLGHEGRGSLHSFLKAKGWATSLSAGVGDDGINRSSLA 394
Query 79 TLLGIKVDLTQKGAAHRGLVLQEIFSYINFLRDHGVGHELVSTMAQQSHIDFHTT--QPS 136
+ G+ + LT G ++ I+ Y+ LRD + + ++DF QP+
Sbjct 395 YVFGMSIHLTDSGLEKIYDIIGYIYQYLKLLRDVSPQEWIFKELQDIGNMDFRFAEEQPA 454
Query 137 SSIMDEAARLAHNLLTYEPYHVVAGDSLLIDADPRLTNQLLQEMSPSKAIIAFSDPDFTS 196
D AA L+ N+L Y HV+ GD + DP+L L+ +P I S
Sbjct 455 D---DYAAELSENMLAYPVEHVIYGDYVYQTWDPKLIEDLMGFFTPQNMRIDVVSKSIKS 511
Query 197 KVDSFETDPYYGVQFRVLDLPQHHAVAMAVLTASPNAFRMPPSLMHIPKASELKILPGLL 256
+ F+ +P++G + D+P + + + N+ +P IP ++ + +
Sbjct 512 --EEFQQEPWFGSSYIEEDVPLSLMESWSNPSEVDNSLHLPSKNQFIPCDFSIRAINSDV 569
Query 257 ---GLTEPELISEQGGNAGTAVWWQGQGAFAIPR----ITVQLNGSILKDKADLLSRTQG 309
+ P I ++ W++ F +PR + L G+ K LL+
Sbjct 570 DPKSQSPPRCIIDE---PFMKFWYKLDETFKVPRANTYFRINLKGAYASVKNCLLTE--- 623
Query 310 SLALAAIAEHLQEETVDFQNCGVTHSLAFKGTGFHM---GFEAYTEGQLSKLMEHVAKLL 366
L + + + L E + +Q + SL+ G + GF LSK++ +AK
Sbjct 624 -LYINLLKDELNE--IIYQATKLETSLSMYGDKLELKVYGFNEKIPALLSKILA-IAKSF 679
Query 367 SDPSMVEPERFERIKQKQMKLVADPATSMA-FEHALEAAAILTRNDAFSRKDLLNALQQT 425
M ERF+ IK+ + + T+M H+ L + + L+ L
Sbjct 680 ----MPNLERFKVIKENMERGFRN--TNMKPLNHSTYLRLQLLCKRIYDSDEKLSVLNDL 733
Query 426 NYDDSFAKLNELKN-VHVDAFVMGNIDRDQSLTMVEEFLEQAGFTPI----DHDDAVASL 480
+ DD + + EL++ + ++A GN+ D+++ + F + P+ H + +
Sbjct 734 SLDDLNSFIPELRSQIFIEALCHGNLSEDEAVNISNIFKDSLTVEPLPSKCRHGEQITCF 793
Query 481 AMEQKQTIEATLANPIKGDKDHASLVQFQLGIPSIEDRVNLAVLTQF---LNRRIYDSLR 537
M K + + N K + + + +Q+ + AVL F + +++ LR
Sbjct 794 PMGAKLVRDVNVKN--KSETNSVVELYYQIEPEEAQSTRTKAVLDLFHEIIEEPLFNQLR 851
Query 538 TEAQLGYIAGAKESQAASTALLQCFVEGAKAHPDEVVKMIDEELSKAKEYLANMPEAEMA 597
T+ QLGY+ V+ +K P ++ +D + + L + +
Sbjct 852 TKEQLGYVVECGPRLTYRVHGFCFCVQSSKYGPVHLLGRVDNFIKDIEGLLEQLDDESYE 911
Query 598 RWKEAAHAKL 607
++ A+L
Sbjct 912 DYRSGMIARL 921
> CE10402
Length=1067
Score = 110 bits (276), Expect = 9e-24, Method: Compositional matrix adjust.
Identities = 135/650 (20%), Positives = 264/650 (40%), Gaps = 47/650 (7%)
Query 24 VAFGLPATLTSYKKQPTSVLTYLLEYTGEGSLAKRLRLLGLADGISPAVDRNSISTLLGI 83
++F P + QP +++L+ + G GSL L+ LG + D ++ + G+
Sbjct 337 ISFPFPDLNGEFLSQPGHYISHLIGHEGPGSLLSELKRLGWVSSLQS--DSHTQAAGFGV 394
Query 84 ---KVDLTQKGAAHRGLVLQEIFSYINFLRDHGVGHELVSTMAQQSHIDFH---TTQPSS 137
+DL+ +G H ++Q +F+YI L+ G + +A+ S + F QP +
Sbjct 395 YNVTMDLSTEGLEHVDEIIQLMFNYIGMLQSAGPKQWVHDELAELSAVKFRFKDKEQPMT 454
Query 138 SIMDEAARLAHNLLTYEPY-HVVAGDSLLIDADPRLTNQLLQEMSPSKAIIAFSDPDFTS 196
++ AA L Y P+ H+++ LL +P +LL +SP+ + F
Sbjct 455 MAINVAASLQ-----YIPFEHILSSRYLLTKYEPERIKELLSMLSPANMQVRVVSQKFKG 509
Query 197 KVDSFETDPYYGVQFRVLDL-PQHHAVAMAVLTASPNAFRMPPSLMHIPKASELKILPGL 255
+ + +P YG + +V D+ P+ L S +A +P +I + K +
Sbjct 510 Q-EGNTNEPVYGTEMKVTDISPETMKKYENALKTSHHALHLPEKNEYIATNFDQKPRESV 568
Query 256 LGLTEPELISEQGGNAGTAVWWQGQGAFAIPRITVQL----------------NGSILKD 299
P LIS+ G + VW++ + +P+ +L + L
Sbjct 569 KN-EHPRLISDDGW---SRVWFKQDDEYNMPKQETKLALTTPMVAQNPRMSLLSSLWLWC 624
Query 300 KADLLSRTQGSLALAAIAEHLQEETVDFQNCGVTHSLAFKGTGFHMGFEAYTEGQLSKLM 359
+D L+ + LA + L+ Q A + + Y E Q +
Sbjct 625 LSDTLAEETYNADLAGLKCQLESSPFGVQMRVSNRREAERHASLTLHVYGYDEKQ-ALFA 683
Query 360 EHVAKLLSDPSMVEPERFERIKQKQMKLVADPATSMAFEHALEAAAILTRNDAFSRKDLL 419
+H+A +++ ++ RF+ + + + + + A S + +L + +S++ LL
Sbjct 684 KHLANRMTN-FKIDKTRFDVLFESLKRALTNHAFSQPYLLTQHYNQLLIVDKVWSKEQLL 742
Query 420 NALQQTNYDD--SFAKLNELKNVHVDAFVMGNIDRDQSLTMVEEFLE-----QAGFTPID 472
+D FAK L+ H++ FV GN +++ + +E ++ P+
Sbjct 743 AVCDSVTLEDVQGFAK-EMLQAFHMELFVHGNSTEKEAIQLSKELMDVLKSAAPNSRPLY 801
Query 473 HDDAVASLAMEQKQTIEATLANPIKGDKDHASLVQFQLGIPSIEDRVNLAVLTQFLNRRI 532
++ ++ E + K V +Q+G+ + D + ++ Q +
Sbjct 802 RNEHNPRRELQLNNGDEYVYRHLQKTHDVGCVEVTYQIGVQNTYDNAVVGLIDQLIREPA 861
Query 533 YDSLRTEAQLGYIAGAKESQAASTALLQCFVEGAKAHPDEVVKMIDEELSKAKEYLANMP 592
+++LRT LGYI T L V+G K+ D V++ I+ L ++ +A MP
Sbjct 862 FNTLRTNEALGYIVWTGSRLNCGTVALNVIVQGPKS-VDHVLERIEVFLESVRKEIAEMP 920
Query 593 EAEMARWKEAAHAKLTKMEANFSEDFKKSAEEIFAHSNCFTKRDLEVKYL 642
+ E A+L + S F++ EI F +R+ EV L
Sbjct 921 QEEFDNQVSGMIARLEEKPKTLSSRFRRFWNEIECRQYNFARREEEVALL 970
> At2g41790
Length=970
Score = 107 bits (267), Expect = 9e-23, Method: Compositional matrix adjust.
Identities = 131/658 (19%), Positives = 268/658 (40%), Gaps = 50/658 (7%)
Query 22 LWVAFGLPATLTSYKKQPTSVLTYLLEYTGEGSLAKRLRLLGLADGISPAVDRNSIS-TL 80
L V++ + ++ Y + P+ L +L+ + GEGSL L+ LG A G+S ++ +
Sbjct 274 LGVSWPVTPSIHHYDEAPSQYLGHLIGHEGEGSLFHALKTLGWATGLSAGEGEWTLDYSF 333
Query 81 LGIKVDLTQKGAAHRGLVLQEIFSYINFLRDHGVGHELVSTMAQQSHIDFHTTQ---PSS 137
+ +DLT G H +L +F+YI L+ GV + ++ FH P S
Sbjct 334 FKVSIDLTDAGHEHMQEILGLLFNYIQLLQQTGVCQWIFDELSAICETKFHYQDKIPPMS 393
Query 138 SIMDEAARLAHNLLTYEPYHVVAGDSLLIDADPRLTNQLLQEMSPSKAIIAFSDPDFTSK 197
I+D +A N+ Y + G SL +P + +++ E+SPS I + F +
Sbjct 394 YIVD----IASNMQIYPTKDWLVGSSLPTKFNPAIVQKVVDELSPSNFRIFWESQKFEGQ 449
Query 198 VDSFETDPYYGVQFRVLDLPQ-------HHAVAMAVLTASPNAFRMPPSLMHIPKASELK 250
D + +P+Y + + + A + + +PN F P+ + + A + +
Sbjct 450 TD--KAEPWYNTAYSLEKITSSTIQEWVQSAPDVHLHLPAPNVFI--PTDLSLKDADDKE 505
Query 251 ILPGLLGLTEPELISEQGGNAGTAVWWQGQGAFAIPRITVQLNGSILKDKADLLSRTQGS 310
+P LL T + +W++ F+ P+ V+++ + + +
Sbjct 506 TVPVLLRKT-----------PFSRLWYKPDTMFSKPKAYVKMDFNCPLAVSSPDAAVLTD 554
Query 311 LALAAIAEHLQEETVDFQNCGVTHSLAFKGTGFHMGFEAYTEGQLSKLMEHVAKLLSDPS 370
+ + ++L E Q G+ + ++ GF + Y +L L+E V +++
Sbjct 555 IFTRLLMDYLNEYAYYAQVAGLYYGVSLSDNGFELTLLGYNH-KLRILLETVVGKIANFE 613
Query 371 MVEPERFERIKQKQMKLVADPATSMAFEHALEAAAILTRNDAFSRKDLLNALQQTNYDDS 430
V+P+RF IK+ K + + A+ +++ ++ + + L+ L +D
Sbjct 614 -VKPDRFAVIKETVTKEYQNYKFRQPYHQAMYYCSLILQDQTWPWTEELDVLSHLEAED- 671
Query 431 FAKLNE--LKNVHVDAFVMGNIDRDQSLTMVEEFLEQAGFT----------PIDH-DDAV 477
AK L ++ ++ GN++ +++ +MV+ +E F P H + V
Sbjct 672 VAKFVPMLLSRTFIECYIAGNVENNEAESMVKH-IEDVLFNDPKPICRPLFPSQHLTNRV 730
Query 478 ASLAMEQKQTIEATLANPIKGDKDHASLVQFQLGIPSIEDRVNLAVLTQFLNRRIYDSLR 537
L K +NP D++ A + Q+ + L + + + LR
Sbjct 731 VKLGEGMKYFYHQDGSNP--SDENSALVHYIQVHRDDFSMNIKLQLFGLVAKQATFHQLR 788
Query 538 TEAQLGYIAGAKESQAASTALLQCFVEGAKAHPDEVVKMIDEELSKAKEYLANMPEAEMA 597
T QLGYI + + +Q ++ + P + ++ L + L M +
Sbjct 789 TVEQLGYITALAQRNDSGIYGVQFIIQSSVKGPGHIDSRVESLLKNFESKLYEMSNEDFK 848
Query 598 RWKEAAHAKLTKMEANFSEDFKKSAEEIFAHSNCFTKRDLEVKYLDNDFSRKQLLRTF 655
A + N E+ + EI + + F +++ EV L +++L+ F
Sbjct 849 SNVTALIDMKLEKHKNLKEESRFYWREIQSGTLKFNRKEAEVSAL-KQLQKQELIDFF 905
> Hs4826770
Length=1019
Score = 105 bits (262), Expect = 4e-22, Method: Compositional matrix adjust.
Identities = 148/695 (21%), Positives = 281/695 (40%), Gaps = 56/695 (8%)
Query 21 SLWVAFGLPATLTSYKKQPTSVLTYLLEYTGEGSLAKRLRLLGLADGISPAVDRNSISTL 80
+L+V F +P YK P L +L+ + G GSL L+ G + + + +
Sbjct 312 NLYVTFPIPDLQKYYKSNPGHYLGHLIGHEGPGSLLSELKSKGWVNTLVGGQKEGARGFM 371
Query 81 LGI-KVDLTQKGAAHRGLVLQEIFSYINFLRDHGVGHELVSTMAQQSHIDFHTTQPSSSI 139
I VDLT++G H ++ +F YI LR G + + + F
Sbjct 372 FFIINVDLTEEGLLHVEDIILHMFQYIQKLRAEGPQEWVFQECKDLNAVAFRFKDKERP- 430
Query 140 MDEAARLAHNLLTYEPYHVVAGDSLLIDADPRLTNQLLQEMSPSKAIIAFSDPDFTSKVD 199
+++A L Y V+ + LL + P L +L ++ P +A F K D
Sbjct 431 RGYTSKIAGILHYYPLEEVLTAEYLLEEFRPDLIEMVLDKLRPENVRVAIVSKSFEGKTD 490
Query 200 SFETDPYYGVQFRVLDLPQHHAVAMAVLTASPNA-----FRMPPSLMHIPKASELKILPG 254
T+ +YG Q++ A+ V+ NA F++P IP + +ILP
Sbjct 491 --RTEEWYGTQYK------QEAIPDEVIKKWQNADLNGKFKLPTKNEFIP--TNFEILPL 540
Query 255 LLGLTE-PELISEQGGNAGTAVWWQGQGAFAIPRITVQLNGSILKDKADLLSRTQGSLAL 313
T P LI + + +W++ P+ + D L L L
Sbjct 541 EKEATPYPALIKD---TVMSKLWFKQDDKKKKPKACLNFEFFSPFAYVDPLHCNMAYLYL 597
Query 314 AAIAEHLQEETVDFQNCGVTHSLAFKGTGFHMGFEAYTEGQ---LSKLMEHVAKLLSDPS 370
+ + L E + G+++ L G ++ + Y + Q L K++E +A
Sbjct 598 ELLKDSLNEYAYAAELAGLSYDLQNTIYGMYLSVKGYNDKQPILLKKIIEKMATF----- 652
Query 371 MVEPERFERIKQKQMKLVADPATSMAFEHALEAAAILTRNDAFSRKDLLNALQQTNYDDS 430
++ +RFE IK+ M+ + + +HA+ +L A+++ +L AL
Sbjct 653 EIDEKRFEIIKEAYMRSLNNFRAEQPHQHAMYYLRLLMTEVAWTKDELKEALDDVTLPRL 712
Query 431 FAKLNEL-KNVHVDAFVMGNIDRDQSL---TMVEEFLEQAGFTPIDHDDAVASLAMEQKQ 486
A + +L +H++A + GNI + +L MVE+ L I+H L + +
Sbjct 713 KAFIPQLLSRLHIEALLHGNITKQAALGIMQMVEDTL-------IEHAHTKPLLPSQLVR 765
Query 487 TIEATLANPIKG-------DKDHASL---VQFQLGIPSIEDRVNLAVLTQFLNRRIYDSL 536
E L P +G ++ H + + +Q + S + + L + Q ++ +++L
Sbjct 766 YREVQL--PDRGWFVYQQRNEVHNNCGIEIYYQTDMQSTSENMFLELFCQIISEPCFNTL 823
Query 537 RTEAQLGYIAGAKESQAASTALLQCFVEGAKAHPDEVVKMIDEELSKAKEYLANMPEAEM 596
RT+ QLGYI + +A L+ ++ K P + ++ L ++ + +M E
Sbjct 824 RTKEQLGYIVFSGPRRANGIQSLRFIIQSEKP-PHYLESRVEAFLITMEKSIEDMTEEAF 882
Query 597 ARWKEAAHAKLTKMEANFSEDFKKSAEEIFAHSNCFTKRDLEVKYLDNDFSRKQLLRTFA 656
+ +A + S + K EI + F + + EV YL +++ +++ +
Sbjct 883 QKHIQALAIRRLDKPKKLSAECAKYWGEIISQQYNFDRDNTEVAYLKT-LTKEDIIKFYK 941
Query 657 KL--SDPSRRMVVKLIADLEPEKEVTLIGEAKAQD 689
++ D RR V + ++GE Q+
Sbjct 942 EMLAVDAPRRHKVSVHVLAREMDSCPVVGEFPCQN 976
> CE06755
Length=980
Score = 105 bits (261), Expect = 5e-22, Method: Compositional matrix adjust.
Identities = 137/640 (21%), Positives = 261/640 (40%), Gaps = 51/640 (7%)
Query 36 KKQPTSVLTYLLEYTGEGSLAKRLRLLGLADGISPAVDRNSIST---LLGIKVDLTQKGA 92
K+ +L+ + G GSL L+ LG + + D N+I+ +L + +DL+ G
Sbjct 263 KRIDRKFFAHLIRHKGPGSLLVELKRLGWVNSLKS--DSNTIAAGFGILNVTMDLSTGGL 320
Query 93 AHRGLVLQEIFSYINFLRDHGVGHELVSTMAQQSHIDFH---TTQPSSSIMDEAARLAHN 149
+ ++Q + +YI L+ G + +A S + F QP ++ AA L
Sbjct 321 ENVDEIIQLMLNYIGMLKSFGPQQWIHDELADLSDVKFRFKDKEQPMKMAINIAASLQ-- 378
Query 150 LLTYEPY-HVVAGDSLLIDADPRLTNQLLQEMSPSKAIIAFSDPDFTSKVDSFETDPYYG 208
Y P H+++ LL +P +LL ++PS ++ F + + +P YG
Sbjct 379 ---YIPIEHILSSRYLLTKYEPERIKELLSTLTPSNMLVRVVSQKFKEQ-EGNTNEPVYG 434
Query 209 VQFRVLDL-PQHHAVAMAVLTASPNAFRMPPSLMHIPKASELKILPGLLGLTE-PELISE 266
+ +V D+ P+ L S +A +P +I A+ P E P+LIS+
Sbjct 435 TEMKVTDISPEKMKKYENALKTSHHALHLPEKNEYI--ATNFGQKPRESVKNEHPKLISD 492
Query 267 QGGNAGTAVWW---------QGQGAFAI--------PRITVQLNGSILKDKADLLSRTQG 309
G + VW+ + + FA+ PRI++ ++ L D+LS
Sbjct 493 DG---WSRVWFKQDDEYNMPKQETKFALTTPIVSQNPRISL-ISSLWLWCFCDILSEETY 548
Query 310 SLALAAIAEHLQEETVDFQNCGVTHSLAFKGTGFHMGFEAYTEGQLSKLMEHVAKLLSDP 369
+ ALA + + Q A + + Y E Q ++H+ + +
Sbjct 549 NAALAGLGCQFELSPFGVQKQSTDGREAERHASLTLHVYGYDEKQ-PLFVKHLTSCMIN- 606
Query 370 SMVEPERFERIKQKQMKLVADPATSMAFEHALEAAAILTRNDAFSRKDLLNALQQTNYDD 429
++ RFE + + + + + A S + +L + +S++ LL ++
Sbjct 607 FKIDRTRFEVLFESLKRTLTNHAFSQPYLLTQHYNQLLIVDKVWSKEQLLAVCDSVTLEN 666
Query 430 --SFAKLNELKNVHVDAFVMGNIDRDQSLTMVEEFLE-----QAGFTPIDHDDAVASLAM 482
FA+ L+ H++ FV GN +++ + +E ++ P+ ++
Sbjct 667 VQGFAR-EMLQAFHMELFVHGNSTEKEAIQLSKELMDILKSAAPNSRPLYRNEHNPRREF 725
Query 483 EQKQTIEATLANPIKGDKDHASLVQFQLGIPSIEDRVNLAVLTQFLNRRIYDSLRTEAQL 542
+ E + K V +Q+G+ + D + ++ Q + ++D+LRT L
Sbjct 726 QLNNGDEYIYRHLQKTHDAGCVEVTYQIGVQNKYDNAVVGLIDQLIKEPVFDTLRTNEAL 785
Query 543 GYIAGAKESQAASTALLQCFVEGAKAHPDEVVKMIDEELSKAKEYLANMPEAEMARWKEA 602
GYI L FV+G K+ D V++ I+ L ++ + MP+ E +
Sbjct 786 GYIVWTGCRFNCGAVALNIFVQGPKS-VDYVLERIEVFLESVRKEIIEMPQDEFEKKVAG 844
Query 603 AHAKLTKMEANFSEDFKKSAEEIFAHSNCFTKRDLEVKYL 642
A+L + S FK+ +I F +R+ EVK L
Sbjct 845 MIARLEEKPKTLSNRFKRFWYQIECRQYDFARREKEVKVL 884
> YLR389c
Length=988
Score = 104 bits (260), Expect = 6e-22, Method: Compositional matrix adjust.
Identities = 131/673 (19%), Positives = 288/673 (42%), Gaps = 35/673 (5%)
Query 24 VAFGLPATLTSYKKQPTSVLTYLLEYTGEGSLAKRLRLLGLADGISPAVDRNSI-STLLG 82
++F +P ++ +P +L++L+ + G GSL L+ LG A+ +S S +
Sbjct 325 ISFTVPDMEEHWESKPPRILSHLIGHEGSGSLLAHLKKLGWANELSAGGHTVSKGNAFFA 384
Query 83 IKVDLTQKGAAHRGLVLQEIFSYINFLRDHGVGHELVSTMAQQSHIDFHTTQ---PSSSI 139
+ +DLT G H V+ IF YI L++ + + + S+ F Q PSS++
Sbjct 385 VDIDLTDNGLTHYRDVIVLIFQYIEMLKNSLPQKWIFNELQDISNATFKFKQAGSPSSTV 444
Query 140 MDEAARLAHNLLTYEPYHVVAGDSLLIDADPRLTNQLLQEMSPSKAIIAFSDPDFTSKVD 199
A L + Y P + LL +P L Q + P + + + ++
Sbjct 445 SSLAKCLEKD---YIPVSRILAMGLLTKYEPDLLTQYTDALVPENSRVTL----ISRSLE 497
Query 200 SFETDPYYGVQFRVLDLPQHHAVAMAVLTASPNAFRMPPSLMHIPKASELKILPGLLGLT 259
+ + +YG ++V+D P M +P A +P + ++ + G+ L
Sbjct 498 TDSAEKWYGTAYKVVDYPADLIKNMKSPGLNP-ALTLPRPNEFVSTNFKVDKIDGIKPLD 556
Query 260 EPELISEQGGNAGTAVWWQGQGAFAIPRITVQLNGSILKDKADLLSRTQGSLALAAIAEH 319
EP L+ + + +W++ F PR + L+ + A +++ +L +
Sbjct 557 EPVLLL---SDDVSKLWYKKDDRFWQPRGYIYLSFKLPHTHASIINSMLSTLYTQLANDA 613
Query 320 LQEETVDFQNCGVTHSLAFKGTGFHMGFEAYTEGQLSKLMEHVAKLLSDPSMVEP--ERF 377
L++ V + ++F T + G G KL+ + + L + EP +RF
Sbjct 614 LKD--VQYDAACADLRISFNKT--NQGLAITASGFNEKLIILLTRFLQGVNSFEPKKDRF 669
Query 378 ERIKQKQMKLVADPATSMAFEHALEAAAILTRNDAFSRKDLLNALQQTNYDDSFAKLNEL 437
E +K K ++ + + + + + ++S + L ++ ++ + +
Sbjct 670 EILKDKTIRHLKNLLYEVPYSQMSNYYNAIINERSWSTAEKLQVFEKLTFEQLINFIPTI 729
Query 438 -KNVHVDAFVMGNIDRDQSL---TMVEEFLEQAGFTPIDHDDAVASLAMEQKQTIEATLA 493
+ V+ + + GNI +++L ++++ + ++ + S + + +T A
Sbjct 730 YEGVYFETLIHGNIKHEEALEVDSLIKSLIPNNIHNLQVSNNRLRSYLLPKGKTFRYETA 789
Query 494 NPIKGDKDHASLVQF--QLGIPSIEDRVNLA-VLTQFLNRRIYDSLRTEAQLGYIAGAKE 550
+K ++ S +Q QL + S ED L+ + Q ++ +D+LRT+ QLGY+ +
Sbjct 790 --LKDSQNVNSCIQHVTQLDVYS-EDLSALSGLFAQLIHEPCFDTLRTKEQLGYVVFSSS 846
Query 551 SQAASTALLQCFVEGAKAHPDEVVKMIDEELSKAKEYLANMPEAEMARWKEAAHAKLTKM 610
TA ++ ++ P + I+ + L +MPE + + KEA L +
Sbjct 847 LNNHGTANIRILIQSEHTTP-YLEWRINNFYETFGQVLRDMPEEDFEKHKEALCNSLLQK 905
Query 611 EANFSEDFKKSAEEIFAHSNCFTKRDLEVKYLDNDFSRKQLLRTFAK--LSDPSRRMVVK 668
N +E+ + I+ FT R + K + N +++Q++ + +S+ + ++++
Sbjct 906 FKNMAEESARYTAAIYLGDYNFTHRQKKAKLVAN-ITKQQMIDFYENYIMSENASKLILH 964
Query 669 LIADLEPEKEVTL 681
L + +E + + +
Sbjct 965 LKSQVETKSSMKM 977
> At3g57470
Length=989
Score = 104 bits (259), Expect = 8e-22, Method: Compositional matrix adjust.
Identities = 124/618 (20%), Positives = 248/618 (40%), Gaps = 68/618 (11%)
Query 24 VAFGLPATLTSYKKQPTSVLTYLLEYTGEGSLAKRLRLLGLADGISPA-VDRNSISTLLG 82
V++ + +++ Y++ P L L+ + GEGSL L++LG A G+ D + +
Sbjct 278 VSWPVTPSISHYEEAPCRYLGDLIGHEGEGSLFHALKILGWATGLYAGEADWSMEYSFFN 337
Query 83 IKVDLTQKGAAHRGLVLQEIFSYINFLRDHGVGHELVSTMAQQSHIDFHTTQPSSSIMDE 142
+ +DLT G H +L +F YI L+ GV + ++ +FH Q +
Sbjct 338 VSIDLTDAGHEHMQDILGLLFEYIKVLQQSGVSQWIFDELSAICEAEFH-YQAKIDPISY 396
Query 143 AARLAHNLLTYEPYHVVAGDSLLIDADPRLTNQLLQEMSPSKA------------IIAFS 190
A ++ N+ Y H + G SL +P + ++L E+SP+ ++
Sbjct 397 AVDISSNMKIYPTKHWLVGSSLPSKFNPAIVQKVLDELSPNNPSVPNVLCSYNLHVVQAL 456
Query 191 DPDFTSKVDSFETDPYYGVQFRVLDLPQ-------HHAVAMAVLTASPNAFRMPPSLMHI 243
+PD + + +P+Y + + + + A + +L +PN F P+ +
Sbjct 457 NPDCLRPRQTDKVEPWYNTAYSLEKITKFTIQEWMQSAPDVNLLLPTPNVFI--PTDFSL 514
Query 244 PKASELKILPGLLGLTEPELISEQGGNAGTAVWWQGQGAFAIPRITVQLNGSILKDKADL 303
+ I P LL T + + +W++ F P+ V+++ + +
Sbjct 515 KDLKDKDIFPVLLRKT-----------SYSRLWYKPDTKFFKPKAYVKMDFNCPLAVSSP 563
Query 304 LSRTQGSLALAAIAEHLQEE---TVDF------QNCGVTHSLAFKGTGFHM---GFEAYT 351
+ + + + ++L E +D+ Q G+ + L+ GF + GF
Sbjct 564 DAAVLSDIFVWLLVDYLNEYALINLDYVSAYYAQAAGLDYGLSLSDNGFELSLAGFNHKL 623
Query 352 EGQLSKLMEHVAKLLSDPSMVEPERFERIKQKQMKLVADPATSMAFEHALEAAAILTRND 411
L +++ +AK V+P+RF IK+ K + E A +++ ++
Sbjct 624 RILLEAVIQKIAKF-----EVKPDRFSVIKETVTKAYQNNKFQQPHEQATNYCSLVLQDQ 678
Query 412 AFSRKDLLNALQQTNYDD--SFAKLNELKNVHVDAFVMGNIDRDQSLTMVEEFLEQAGFT 469
+ + L+AL +D +F + L V+ ++ GN+++D++ +MV+ +E FT
Sbjct 679 IWPWTEELDALSHLEAEDLANFVPM-LLSRTFVECYIAGNVEKDEAESMVKH-IEDVLFT 736
Query 470 ---PIDH--------DDAVASLAMEQKQTIEATLANPIKGDKDHASLVQFQLGIPSIEDR 518
PI + V L K +N D++ A + Q+
Sbjct 737 DSKPICRPLFPSQFLTNRVTELGTGMKHFYYQEGSN--SSDENSALVHYIQVHKDEFSMN 794
Query 519 VNLAVLTQFLNRRIYDSLRTEAQLGYIAGAKESQAASTALLQCFVEGAKAHPDEVVKMID 578
L + + + LRT QLGYI S + +Q ++ + P + ++
Sbjct 795 SKLQLFELIAKQDTFHQLRTIEQLGYITSLSLSNDSGVYGVQFIIQSSVKGPGHIDSRVE 854
Query 579 EELSKAKEYLANMPEAEM 596
L + NM + E
Sbjct 855 SLLKDLESKFYNMSDEEF 872
> CE06756
Length=845
Score = 100 bits (248), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 118/568 (20%), Positives = 237/568 (41%), Gaps = 41/568 (7%)
Query 24 VAFGLPATLTSYKKQPTSVLTYLLEYTGEGSLAKRLRLLGLADGISPAVDRNSISTLLG- 82
+ F P Y QP + +L+ + G GS++ L+ LG A + P + +I+ G
Sbjct 278 IIFPFPDLNNEYLSQPGHYIAHLIGHKGPGSISSELKRLGWASSLKP--ESKTIAAGFGY 335
Query 83 --IKVDLTQKGAAHRGLVLQEIFSYINFLRDHGVGHELVSTMAQQSHIDFHTTQPSSSIM 140
+ +DL+ +G H ++Q +F+YI L+ G + +A+ S I+F + +
Sbjct 336 FNVTMDLSTEGLEHVDEIIQLMFNYIGMLQSAGPQQWIHEELAELSAIEFR-FKDREPLT 394
Query 141 DEAARLAHNLLTYEPY-HVVAGDSLLIDADPRLTNQLLQEMSPSKAIIAFSDPDFTSKVD 199
A ++A N L Y P+ H+++ LL +P +LL ++PS ++ F + +
Sbjct 395 KNAIKVARN-LQYIPFEHILSSRYLLTKYNPERIKELLSTLTPSNMLVRVVSKKFKEQ-E 452
Query 200 SFETDPYYGVQFRVLDL-PQHHAVAMAVLTASPNAFRMPPSLMHIPKASELKILPGLLGL 258
+P YG + +V D+ P+ L S +A +P +I + K +
Sbjct 453 GNTNEPVYGTEMKVTDISPEKMKKYENALKTSHHALHLPEKNEYIVTKFDQKPRESVKN- 511
Query 259 TEPELISEQGGNAGTAVWWQGQGAFAIPRITVQLNGSILKDKADLLSRTQGSLALAAIAE 318
P LIS+ G + VW++ + +P+ +L + + + SL L + +
Sbjct 512 EHPRLISDDG---WSRVWFKQDDEYNMPKQETKLAFTTPIVAQNPIMSLISSLWLWCLND 568
Query 319 HLQEETVDFQNCGVTHSLAFKGTGFH----------------MGFEAYTEGQLSKLMEHV 362
L EET + G+ L G H + Y E Q ++H+
Sbjct 569 TLTEETYNAAIAGLKFQLESGHNGVHEQAGNWLDPERHASITLHVYGYDEKQ-PLFVKHL 627
Query 363 AKLLSDPSMVEPERFERIKQKQMKLVADPATSMAFEHALEAAAILTRNDAFSRKDLLNAL 422
K +++ ++ RF+ + + + + + A S + + +L +S++ LL
Sbjct 628 TKCMTN-FKIDRTRFDVVFESLKRSLTNHAFSQPYMLSKYFNELLVVEKVWSKEQLLAVC 686
Query 423 QQTNYDDSFAKLNEL-KNVHVDAFVMGNIDRDQSLTMVEEFLE----QAGFTPIDHDDAV 477
+D EL + H++ FV GN +++ + E ++ A + + + +
Sbjct 687 DSATLEDVQGFSKELFQAFHLELFVHGNSTEKKAIQLSNELMDILKSAAPNSRLLYRNEH 746
Query 478 ASLAMEQKQTIEATLANPIKGDKDHASL-VQFQLGIPSIEDRVNLAVLTQFLNRRIYDSL 536
Q + + ++ D + V F+ G+ + D +++Q + + + +L
Sbjct 747 NPRREFQLNNGDEYIYRHLQKTHDAGCVEVTFKFGVQNTYDNALAGLISQLIRQPAFSTL 806
Query 537 RTEAQLG---YIAGAKESQAASTALLQC 561
RT+ LG Y K S+ LL+C
Sbjct 807 RTKESLGKNSYPHLIKYSKYLERKLLRC 834
> SPACUNK4.12c
Length=969
Score = 97.8 bits (242), Expect = 8e-20, Method: Compositional matrix adjust.
Identities = 132/600 (22%), Positives = 246/600 (41%), Gaps = 53/600 (8%)
Query 24 VAFGLPATLTSYKKQPTSVLTYLLEYTGEGSLAKRLRLLGLADG-ISPAVDRNSISTLLG 82
+ F +P YK +P + +LL + GEGS L+ LGLA I+ V + ++
Sbjct 275 IVFPIPGQYHKYKCRPAEYVCHLLGHEGEGSYLAYLKSLGLATSLIAFNVSITEDADIIV 334
Query 83 IKVDLTQKGAAHRGLVLQEIFSYINFLRDHGVGHELVSTMAQQSHIDFHTTQPSSSIMDE 142
+ LT++G V++ +F YI L L S F T Q +
Sbjct 335 VSTFLTEEGLTDYQRVIKILFEYIRLLDQTNAHKFLFEETRIMSEAQFKTRQKTP----- 389
Query 143 AARLAHNLLT-----YEPYHVVAGDSLLIDADPRLTNQLLQEMSPSKAIIAFSDPDFTSK 197
A + AH + + Y V+ S+L + DP+ ++++ + P+ +
Sbjct 390 AYQYAHVVASKLQREYPRDKVLYYSSVLTEFDPKGIQEVVESLRPNNFFAILAAHSIEKG 449
Query 198 VDSFETDPYYGVQFRVLDLPQHHAVAMAVLTASPNAFRMPPSLMHIPKASELKILPGLLG 257
+D+ E +YG+ + + DL ++ + S + +P + IP + E++ P
Sbjct 450 LDNKEK--FYGIDYGLEDLDSQFIDSLLHIKTSSELY-LPLANEFIPWSLEVEKQPVTTK 506
Query 258 LTEPELISEQGGNAGTAVWWQGQGAFAIPRITVQLNG-SILKDKADLLSRTQGSLALAAI 316
L P L+ + +W + F +P+ V +N S + ++ +S +L I
Sbjct 507 LKVPNLVR---NDKFVRLWHKKDDTFWVPKANVFINFISPIARRSPKVS-VSTTLYTRLI 562
Query 317 AEHLQEETVDFQNCGVTHSLAFKGTGFHMGFEAYTEGQLSKLMEHVAKLLSDPSMVEPER 376
+ L E + G++ SL+ G + +T+ +L L+E V ++ D V P+R
Sbjct 563 EDALGEYSYPASLAGLSFSLSPSTRGIILCISGFTD-KLHVLLEKVVAMMRDLK-VHPQR 620
Query 377 FERIKQKQMKLVADPATSMAFEHALEAAAILTRNDAFSRKDLLNALQQTNYDDSFAKLNE 436
FE +K + + + D A+ + L+ ++S +L A++ D +++
Sbjct 621 FEILKNRLEQELKDYDALEAYHRSNHVLTWLSEPHSWSNAELREAIKDVQVGDMSDFISD 680
Query 437 -LKNVHVDAFVMGNIDRDQSLTMVEEFLEQAGFTPIDHDDAVASLAMEQKQTIEATLAN- 494
LK +++ V GN + + ++E + ID AS ++ I N
Sbjct 681 LLKQNFLESLVHGNYTEEDAKNLIE-----SAQKLIDPKPVFASQLSRKRAIIVPEGGNY 735
Query 495 ------PIKGDKDHASLVQFQLGIPSIEDRVNLAVL---TQFLNRRIYDSLRTEAQLGYI 545
P K +K+ A + + L I ++D + A+ Q + + LRT+ QLGYI
Sbjct 736 IYKTVVPNKEEKNSA--IMYNLQISQLDDERSGALTRLARQIMKEPTFSILRTKEQLGYI 793
Query 546 AGAKESQAASTALLQCFVEGAKA--------------HPDEVVKMIDEELSKAKEYLANM 591
Q L FV+ ++ E ++M DE+ SK K L N
Sbjct 794 VFTLVRQVTPFINLNIFVQSERSSTYLESRIRALLDQFKSEFLEMSDEDFSKHKSSLINF 853
> Hs4505453
Length=1219
Score = 94.7 bits (234), Expect = 7e-19, Method: Compositional matrix adjust.
Identities = 141/675 (20%), Positives = 280/675 (41%), Gaps = 59/675 (8%)
Query 21 SLWVAFGLPATLTSYKKQPTSVLTYLLEYTGEGS----LAKRLRLLGLADGIS-PAVDRN 75
+L + + LP Y+ +P +++L+ + G+GS L K+ L L G ++N
Sbjct 507 ALTITWALPPQQQHYRVKPLHYISWLVGHEGKGSILSFLRKKCWALALFGGNGETGFEQN 566
Query 76 SISTLLGIKVDLTQKGAAHRGLVLQEIFSYINFLRDHGVGHELVSTMAQQSHIDFHTTQP 135
S ++ I + LT +G H V +F Y+ L+ G + + + +FH Q
Sbjct 567 STYSVFSISITLTDEGYEHFYEVAYTVFLYLKMLQKLGPEKRIFEEIRKIEDNEFHY-QE 625
Query 136 SSSIMDEAARLAHNLLTYEPYHVVAGDSLLIDADPRLTNQLLQEMSPSKAIIAFSDPDFT 195
+ ++ + N+ Y ++ GD LL + P + + L ++ P KA +
Sbjct 626 QTDPVEYVENMCENMQLYPLQDILTGDQLLFEYKPEVIGEALNQLVPQKANLVLLSGANE 685
Query 196 SKVDSFETDPYYGVQFRVLDLPQHHAVAMAVLTASPNAFRMPPSLMHIPKASELKILPGL 255
K D E ++G Q+ + D+ + A L S F + P L H+P +E K +
Sbjct 686 GKCDLKEK--WFGTQYSIEDIEN----SWAELWNS--NFELNPDL-HLP--AENKYIATD 734
Query 256 LGLT-----EPELISEQGGNAGTAVWWQGQGAFAIPRITVQ---LNGSILKDKADL-LSR 306
L E E + +W++ F IP+ ++ ++ I K A++ L
Sbjct 735 FTLKAFDCPETEYPVKIVNTPQGCLWYKKDNKFKIPKAYIRFHLISPLIQKSAANVVLFD 794
Query 307 TQGSLALAAIAEHLQEETV---DFQNCGVTHSLAFKGTGFHMGFEAYTEGQLSKLMEHVA 363
++ +AE E V +++ H L + GF+ +++++A
Sbjct 795 IFVNILTHNLAEPAYEADVAQLEYKLAAGEHGLIIRVKGFNHKLPLL----FQLIIDYLA 850
Query 364 KLLSDPSMVEPERFERIKQKQMKLVADPATSMAFEHALEAAAILTRNDAFSRKDLLNALQ 423
+ S P++ E++K+ ++ P T A + ++ +S D AL
Sbjct 851 EFNSTPAVFTMIT-EQLKKTYFNILIKPETL-----AKDVRLLILEYARWSMIDKYQALM 904
Query 424 QT-NYDDSFAKLNELKN-VHVDAFVMGNIDRDQSLTMVEEFLEQAGFTPIDHDDAVASLA 481
+ + + + E K+ + V+ V GN+ +S+ ++ +++ F P++ + V
Sbjct 905 DGLSLESLLSFVKEFKSQLFVEGLVQGNVTSTESMDFLKYVVDKLNFKPLEQEMPV---- 960
Query 482 MEQKQTIEATLANPI-------KGDKDHASLVQFQLGIPSIEDRVNLAVLTQFLNRRIYD 534
Q Q +E + + KGD + V +Q G S+ + + +L + +D
Sbjct 961 --QFQVVELPSGHHLCKVKALNKGDANSEVTVYYQSGTRSLREYTLMELLVMHMEEPCFD 1018
Query 535 SLRTEAQLGY--IAGAKESQAASTALLQCFVEGAKAHPDEVVKMIDEELSKAKEYLANMP 592
LRT+ LGY + + + + K + + V K I+E LS +E + N+
Sbjct 1019 FLRTKQTLGYHVYPTCRNTSGILGFSVTVGTQATKYNSEVVDKKIEEFLSSFEEKIENLT 1078
Query 593 EAEMARWKEAAHAKLTKME-ANFSEDFKKSAEEIFAHSNCFTKRDLEVKYLDNDFSRKQL 651
E E + A KL + E + E+ ++ E+ F + E++ L + FS+ L
Sbjct 1079 E-EAFNTQVTALIKLKECEDTHLGEEVDRNWNEVVTQQYLFDRLAHEIEALKS-FSKSDL 1136
Query 652 LRTFAKLSDPSRRMV 666
+ F P +M+
Sbjct 1137 VNWFKAHRGPGSKML 1151
> 7292708
Length=1077
Score = 80.1 bits (196), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 136/663 (20%), Positives = 262/663 (39%), Gaps = 69/663 (10%)
Query 18 SEPSLWVAFGLPATLTSYKKQPTSVLTYLLEYTGEGSLA----KRLRLLGLADGISP-AV 72
+E L + + LP Y+ +P L+YLL Y G GSL +RL L L GI
Sbjct 319 NETKLELTWVLPNVRQYYRSKPDQFLSYLLGYEGRGSLCAYLRRRLWALQLIAGIDENGF 378
Query 73 DRNSISTLLGIKVDLTQKGAAHRGLVLQEIFSYINFLRDHGVGHELVSTMA--QQSHIDF 130
D NS+ +L I + LT +G + VL F+Y+ + G ++ +++ F
Sbjct 379 DMNSMYSLFNICIYLTDEGFKNLDEVLAATFAYVKLFANCGSMKDVYEEQQRNEETGFRF 438
Query 131 HTTQPSSSIMDEAARLAHNLLTYEPYHVVAGDSLLIDADPRLTNQL---LQEMSPSKAII 187
H +P+ D L NL + P ++ G L + + +L L EM + +
Sbjct 439 HAQRPA---FDNVQELVLNLKYFPPKDILTGKELYYEYNEEHLKELISHLNEMKFNLMVT 495
Query 188 AFSDPDFTSKVDSFETDPYYGVQFRVLDLPQHHAVAMAVLTASPNAFRMPPSLMHIPKAS 247
+ D S D +T+ ++G ++ + +P+ P F +P S ++
Sbjct 496 SRRKYDDISAYD--KTEEWFGTEYATIPMPEKWRKLWEDSVPLPELF-LPESNKYV--TD 550
Query 248 ELKILPGLLGLTE----PELISEQGGNAGTAVWWQGQGAFAIPRITVQLNGSILKDKADL 303
+ + +G E P+L+ + +W++ F +P + + +
Sbjct 551 DFTLHWHSMGRPEVPDSPKLLIKTD---TCELWFRQDDKFDLPEAHMAFYFISPMQRQNA 607
Query 304 LSRTQGSLALAAIAEHLQEETVDFQNCGVTHSLAFKGTGFHMGFEAYTEGQLSKLMEHVA 363
+ SL + H+ EE + G+++SL+ G + Y E +L ++E +A
Sbjct 608 KNDAMCSLYEEMVRFHVCEELYPAISAGLSYSLSTIEKGLLLKVCGYNE-KLHLIVEAIA 666
Query 364 K-LLSDPSMVEPERFERIKQKQMKLVADPATSMAFEHALEAAAILTRNDAFSRKDLLNAL 422
+ +L+ ++ + Q K AF +AL L R+ + + L
Sbjct 667 EGMLNVAETLDENMLSAFVKNQRK---------AFFNALIKPKALNRDIRLCVLERIRWL 717
Query 423 QQTNYDD-SFAKLNEL--------KNVHVDAFVMGNIDRDQSLTMVEEFLEQAGFTPIDH 473
Y S L ++ K +++ + + GN + + ++ L + I
Sbjct 718 MINKYKCLSSVILEDMREFAHQFPKELYIQSLIQGNYTEESAHNVMNSLLSRLNCKQI-- 775
Query 474 DDAVASLAMEQKQTIE-ATLANPIKGD--KDHASLVQ---------FQLGIPSIEDRVNL 521
E+ + +E T+ P+ + HA VQ +Q+G ++ L
Sbjct 776 --------RERGRFLEDITVKLPVGTSIIRCHALNVQDTNTVITNFYQIGPNTVRVESIL 827
Query 522 AVLTQFLNRRIYDSLRTEAQLGYIAGAKESQAASTALLQCFVEG--AKAHPDEVVKMIDE 579
+L F++ ++D LRT+ QLGY GA A V K D V I+
Sbjct 828 DLLMMFVDEPLFDQLRTKEQLGYHVGATVRLNYGIAGYSIMVNSQETKTTADYVEGRIEV 887
Query 580 ELSKAKEYLANMPEAEMARWKEAAHAKLTKMEANFSEDFKKSAEEIFAHSNCFTKRDLEV 639
+K + L ++P+ E +++ + S + ++ +EI S F +R ++
Sbjct 888 FRAKMLQILRHLPQDEYEHTRDSLIKLKLVADLALSTEMSRNWDEIINESYLFDRRRRQI 947
Query 640 KYL 642
+ +
Sbjct 948 EVI 950
> CE16673
Length=301
Score = 57.4 bits (137), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 37/137 (27%), Positives = 65/137 (47%), Gaps = 1/137 (0%)
Query 506 VQFQLGIPSIEDRVNLAVLTQFLNRRIYDSLRTEAQLGYIAGAKESQAASTALLQCFVEG 565
V +Q+G+ + D + ++ + +D+LRT+ LGYI + T LQ V+G
Sbjct 76 VTYQIGVQNTYDNAVIGLIKNLITEPAFDTLRTKESLGYIVWTRTHFNCGTVALQILVQG 135
Query 566 AKAHPDEVVKMIDEELSKAKEYLANMPEAEMARWKEAAHAKLTKMEANFSEDFKKSAEEI 625
K+ D V++ I+ L ++ + MP+ E A+L + S FKK +EI
Sbjct 136 PKS-VDHVLERIEAFLESVRKEIVEMPQEEFENRVSGLIAQLEEKPKTLSCRFKKFWDEI 194
Query 626 FAHSNCFTKRDLEVKYL 642
FT+ + +V+ L
Sbjct 195 ECRQYNFTRIEEDVELL 211
> CE01818
Length=745
Score = 51.6 bits (122), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 37/129 (28%), Positives = 65/129 (50%), Gaps = 9/129 (6%)
Query 24 VAFGLPATLTSYKKQPTSVLTYLLEYTGEGSLAKRLRLLGLADGISPAVDRNSISTLLGI 83
+ F P + QP + +L+ + G GSL L+ LG IS D ++I++ G+
Sbjct 279 IKFPFPDLNGEFLSQPGDYIAHLIGHEGPGSLLSELKRLGWV--ISLEADNHTIASGFGV 336
Query 84 ---KVDLTQKGAAHRGLVLQEIFSYINFLRDHGVGHELVSTMAQQSHIDFHTTQPSSSIM 140
+DL+ +G H V+Q +F++I FL+ G + +A+ + +DF +I+
Sbjct 337 FSVTMDLSTEGLEHVDDVIQLVFNFIGFLKSSGPQKWIHDELAELNAVDFRFD--DDTII 394
Query 141 DEA--ARLA 147
+E ARLA
Sbjct 395 EETYNARLA 403
Score = 41.2 bits (95), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 60/276 (21%), Positives = 111/276 (40%), Gaps = 17/276 (6%)
Query 393 TSMAFE--HALEAAAI--LTRNDAFSRKDLLNALQQTNYDD--SFAKLNELKNVHVDAFV 446
T+ AF H L A I L ++ +S++ LL +D FA + L+ H++ FV
Sbjct 460 TNHAFSQPHDLSAHFIDLLVVDNIWSKEQLLAVCDSVTLEDVHGFA-IKMLQAFHMELFV 518
Query 447 MGNIDRDQSLTMVEEFLE-----QAGFTPIDHDDAVASLAMEQKQTIEATLANPIKGDKD 501
GN +L + +E + P+ D+ ++ E + K
Sbjct 519 HGNSTEKDTLQLSKELSDILKSVAPNSRPLKRDEHNPHRELQLINGHEHVYRHFQKTHDV 578
Query 502 HASLVQFQLGIPSIEDRVNLAVLTQFLNRRIYDSLRTEAQLGYIAGAKESQAASTALLQC 561
V FQ+G+ S + +L + + Y LRT LGY + L
Sbjct 579 GCVEVAFQIGVQSTYNNSVNKLLNELIKNPAYTILRTNEALGYNVSTESRLNDGNVYLHV 638
Query 562 FVEGAKAHPDEVVKMIDEELSKAKEYLANMPEAEMARWKEAAHAKLTKMEANFSEDFKKS 621
V+G ++ D V++ I+ L A+E + MP+ + + A + S+ F
Sbjct 639 IVQGPES-ADHVLERIEVFLESAREEIVAMPQED---FDYQVWAMFKENPPTLSQCFSMF 694
Query 622 AEEIFAHSNCFTKRDLEVKYLDNDFSRKQLLRTFAK 657
EI + F R+ EV+ + ++++++ F +
Sbjct 695 WSEIHSRQYNFG-RNKEVRGISKRITKEEVINFFDR 729
> 7296373
Length=908
Score = 43.5 bits (101), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 27/92 (29%), Positives = 45/92 (48%), Gaps = 8/92 (8%)
Query 23 WVAFGLPATLTSYKKQPTSVLTYLLEYTGEGSLAKRLR----LLGLADGIS-PAVDRNSI 77
WV LP Y+ +P ++ L+ Y G GSL LR + + G++ + D NSI
Sbjct 327 WV---LPPMKNFYRSKPDMFISQLIGYEGVGSLCAYLRHHLWCISVVAGVAESSFDSNSI 383
Query 78 STLLGIKVDLTQKGAAHRGLVLQEIFSYINFL 109
+L I + L+ G H VL+ F+++ +
Sbjct 384 YSLFNICIYLSDDGFDHIDEVLEATFAWVKLI 415
Score = 42.7 bits (99), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 67/324 (20%), Positives = 130/324 (40%), Gaps = 23/324 (7%)
Query 331 GVTHSLAFKGTGFHMGFEAYTEGQLSKLMEHVAKLLS--DPSMVEPERFERIKQKQMKLV 388
G+T+ L G M Y E +L L+E + ++ + + + F+ +K++Q+
Sbjct 484 GLTYGLYIGDKGLVMRVSGYNE-KLPLLVEIILNMMQTIELDIGQVNAFKDLKKRQIYNA 542
Query 389 ADPATSMAFEHALEAAAILTRNDAFSRKDLLNALQQTNYDD--SFAKLNELKNVHVDAFV 446
S+ + L + N FS ++ DD SF K N K ++V +
Sbjct 543 LINGKSLNLDLRLS----ILENKRFSMISKYESVDDITMDDIKSF-KENFHKKMYVKGLI 597
Query 447 MGNIDRDQSLTMVEEFLEQAGFTPIDHDDAVASLAMEQKQTIEATLANPIKGDKDHASLV 506
GN DQ+ ++++ L+ +D+ A+ + ++ A + D + +
Sbjct 598 QGNFTEDQATDLMQKVLDTYKSEKLDNLSALDNHLLQIPLGSYYLRAKTLNEDDSNTIIT 657
Query 507 QF-QLG-----IPSIEDRVNLAVLTQFLNRRIYDSLRTEAQLGYIAGAKESQAASTA--L 558
+ Q+G + I D V L V F N+ LRT+ QLGY G + L
Sbjct 658 NYYQIGPSDLKMECIMDLVELIVEEPFFNQ-----LRTQEQLGYSLGIHQRIGYGVLAFL 712
Query 559 LQCFVEGAKAHPDEVVKMIDEELSKAKEYLANMPEAEMARWKEAAHAKLTKMEANFSEDF 618
+ + K D V + I+ S+ E ++ M + E +E + + E+
Sbjct 713 ITINTQETKHRADYVEQRIEAFRSRMAELVSQMSDTEFKNIRETLINGKKLGDTSLDEEV 772
Query 619 KKSAEEIFAHSNCFTKRDLEVKYL 642
++ EI F + + +++ L
Sbjct 773 LRNWSEIVTKEYFFNRIETQIQML 796
> At3g22150
Length=820
Score = 33.5 bits (75), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 48/223 (21%), Positives = 86/223 (38%), Gaps = 28/223 (12%)
Query 428 DDSFAKLNELKNVHVDAFVMGNIDRDQSLTMVEEFL--EQAGFTPIDHDDAVASLAMEQK 485
+D F++ E +V ++G + FL +++G P DA+ +A+
Sbjct 576 EDMFSQTKERNSVTYTTMILGYGQHGMGERAISLFLSMQESGIKP----DAITFVAVLSA 631
Query 486 QTIEATLANPIKGDKDHASLVQFQLGIPSIEDRVNLAVLTQFLNRRIYDSLRTEAQLGYI 545
+ + +K ++ + Q PS E + +T L R R ++
Sbjct 632 CSYSGLIDEGLKIFEEMREVYNIQ---PSSE---HYCCITDMLGR----VGRVNEAYEFV 681
Query 546 AGAKESQAASTALLQCFVEGAKAHPD-EVVKMIDEELSK---AKEY------LANMPEAE 595
G E + L + K H + E+ + + E L+K K + L+NM AE
Sbjct 682 KGLGE-EGNIAELWGSLLGSCKLHGELELAETVSERLAKFDKGKNFSGYEVLLSNM-YAE 739
Query 596 MARWKEAAHAKLTKMEANFSEDFKKSAEEIFAHSNCFTKRDLE 638
+WK + E ++ +S EI + NCF RD E
Sbjct 740 EQKWKSVDKVRRGMREKGLKKEVGRSGIEIAGYVNCFVSRDQE 782
> At5g18240
Length=402
Score = 32.7 bits (73), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 26/83 (31%), Positives = 40/83 (48%), Gaps = 14/83 (16%)
Query 313 LAAIAEHLQEETVDFQNCGVTHSLAFKGTGFHMGFEAYTEGQLSKLMEHVAKLLSDPSMV 372
L +I E QE T+ QN G G EA T+ QLS+L+ V+ D S +
Sbjct 182 LQSILEKAQE-TLGRQNLGAA------------GIEA-TKAQLSELVSKVSADYPDSSFL 227
Query 373 EPERFERIKQKQMKLVADPATSM 395
EP+ + + +QM+ P +S+
Sbjct 228 EPKELQNLHHQQMQKTYPPNSSL 250
> Hs20538161
Length=437
Score = 31.2 bits (69), Expect = 9.2, Method: Compositional matrix adjust.
Identities = 25/92 (27%), Positives = 42/92 (45%), Gaps = 17/92 (18%)
Query 498 GDKDHASLVQFQLGIPSIEDR---------VNLAVLTQFLNRRIYDSLRTEAQLGYIAGA 548
GD+D++ +Q +P + + +NL + + LN + + TEAQ G ++
Sbjct 284 GDRDYSGKDYYQHNLPFVSENHTLSLVVIVMNLIISSSMLNMAMIGKIHTEAQKGRLSSQ 343
Query 549 KESQAA------STALLQCFVEGAKAHPDEVV 574
E Q+ +T +C V GAK HP V
Sbjct 344 PEIQSGLVQLRITTTPCKC-VHGAK-HPQVFV 373
Lambda K H
0.317 0.131 0.369
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 21776317878
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40