bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_1215_orf1
Length=117
Score E
Sequences producing significant alignments: (Bits) Value
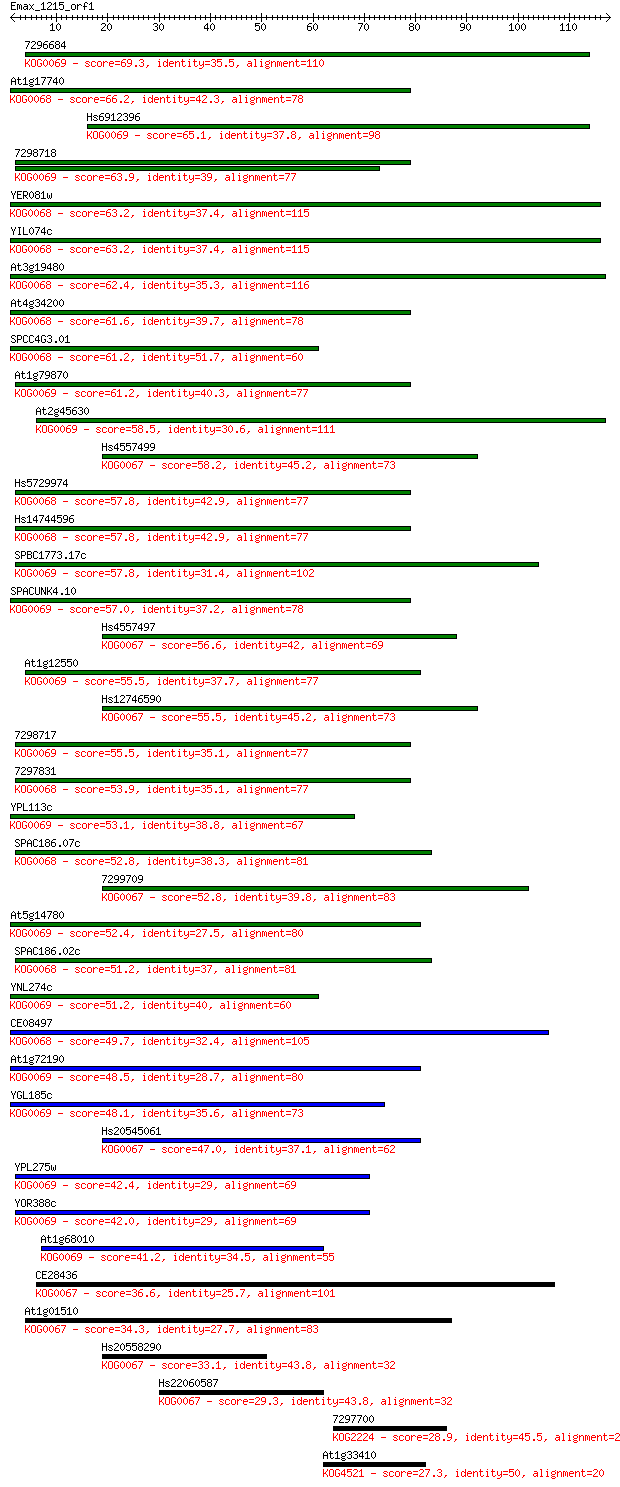
7296684 69.3 2e-12
At1g17740 66.2 1e-11
Hs6912396 65.1 3e-11
7298718 63.9 7e-11
YER081w 63.2 1e-10
YIL074c 63.2 1e-10
At3g19480 62.4 2e-10
At4g34200 61.6 4e-10
SPCC4G3.01 61.2 4e-10
At1g79870 61.2 5e-10
At2g45630 58.5 3e-09
Hs4557499 58.2 4e-09
Hs5729974 57.8 5e-09
Hs14744596 57.8 5e-09
SPBC1773.17c 57.8 5e-09
SPACUNK4.10 57.0 8e-09
Hs4557497 56.6 1e-08
At1g12550 55.5 2e-08
Hs12746590 55.5 2e-08
7298717 55.5 3e-08
7297831 53.9 7e-08
YPL113c 53.1 1e-07
SPAC186.07c 52.8 2e-07
7299709 52.8 2e-07
At5g14780 52.4 2e-07
SPAC186.02c 51.2 5e-07
YNL274c 51.2 5e-07
CE08497 49.7 1e-06
At1g72190 48.5 3e-06
YGL185c 48.1 4e-06
Hs20545061 47.0 9e-06
YPL275w 42.4 2e-04
YOR388c 42.0 3e-04
At1g68010 41.2 4e-04
CE28436 36.6 0.013
At1g01510 34.3 0.051
Hs20558290 33.1 0.12
Hs22060587 29.3 1.9
7297700 28.9 2.5
At1g33410 27.3 6.4
> 7296684
Length=325
Score = 69.3 bits (168), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 39/110 (35%), Positives = 58/110 (52%), Gaps = 1/110 (0%)
Query 4 TAETKQCISYSQFNAMKKNAVFISIGRGSVVDEEALLHALHSGQIAGAALDVFNKEPLPV 63
T ETK+ + + F MK N + I+ RG VVD++AL AL + +I A LDV EPLP+
Sbjct 216 TPETKEIFNATAFQKMKPNCILINTARGGVVDQKALYEALKTKRILAAGLDVTTPEPLPI 275
Query 64 DSPLWSSPNLLLSSHNADFCDNFSAAATLRVFNANMIKYLRGASSNADME 113
D PL N+++ H D + R+ N++ L G A++E
Sbjct 276 DDPLLKLDNVVILPHIGS-ADIETRKEMSRITARNILAALAGDKMVAEVE 324
> At1g17740
Length=624
Score = 66.2 bits (160), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 33/78 (42%), Positives = 48/78 (61%), Gaps = 0/78 (0%)
Query 1 LPATAETKQCISYSQFNAMKKNAVFISIGRGSVVDEEALLHALHSGQIAGAALDVFNKEP 60
+P T TK+ + F+ MKK I++ RG V+DE+AL+ AL +G +A AALDVF +EP
Sbjct 283 MPLTPATKKVFNDETFSKMKKGVRLINVARGGVIDEDALVRALDAGIVAQAALDVFCEEP 342
Query 61 LPVDSPLWSSPNLLLSSH 78
DS L N+ ++ H
Sbjct 343 PSKDSRLIQHENVTVTPH 360
> Hs6912396
Length=328
Score = 65.1 bits (157), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 37/98 (37%), Positives = 55/98 (56%), Gaps = 1/98 (1%)
Query 16 FNAMKKNAVFISIGRGSVVDEEALLHALHSGQIAGAALDVFNKEPLPVDSPLWSSPNLLL 75
F MK+ AVFI+I RG VV+++ L AL SG+IA A LDV + EPLP + PL + N ++
Sbjct 231 FQKMKETAVFINISRGDVVNQDDLYQALASGKIAAAGLDVTSPEPLPTNHPLLTLKNCVI 290
Query 76 SSHNADFCDNFSAAATLRVFNANMIKYLRGASSNADME 113
H +L N N++ LRG ++++
Sbjct 291 LPHIGSATHRTRNTMSLLAAN-NLLAGLRGEPMPSELK 327
> 7298718
Length=598
Score = 63.9 bits (154), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 30/77 (38%), Positives = 48/77 (62%), Gaps = 0/77 (0%)
Query 2 PATAETKQCISYSQFNAMKKNAVFISIGRGSVVDEEALLHALHSGQIAGAALDVFNKEPL 61
P T ET + + FN MK+ AV +++GRG +V+++ L AL S +I A LDV + EPL
Sbjct 488 PLTKETLGLFNATVFNKMKETAVLVNVGRGKIVNQDDLYEALKSNRIFAAGLDVMDPEPL 547
Query 62 PVDSPLWSSPNLLLSSH 78
P + L + N++++ H
Sbjct 548 PSNDKLLTLDNVVVTPH 564
Score = 43.9 bits (102), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 24/71 (33%), Positives = 35/71 (49%), Gaps = 20/71 (28%)
Query 2 PATAETKQCISYSQFNAMKKNAVFISIGRGSVVDEEALLHALHSGQIAGAALDVFNKEPL 61
P T ET++ + FN MK+++VF+++ RG LDV EPL
Sbjct 216 PLTNETREKFNGKAFNLMKRSSVFVNVARG--------------------GLDVTTPEPL 255
Query 62 PVDSPLWSSPN 72
P +SPL + PN
Sbjct 256 PANSPLLNVPN 266
> YER081w
Length=469
Score = 63.2 bits (152), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 43/127 (33%), Positives = 66/127 (51%), Gaps = 13/127 (10%)
Query 1 LPATAETKQCISYSQFNAMKKNAVFISIGRGSVVDEEALLHALHSGQIAGAALDVFNKEP 60
+PAT ET++ +S QF AMK A I+ RG+VVD +L+ A+ + +IAGAALDV+ EP
Sbjct 258 VPATPETEKMLSAPQFAAMKDGAYVINASRGTVVDIPSLIQAVKANKIAGAALDVYPHEP 317
Query 61 LP------------VDSPLWSSPNLLLSSHNADFCDNFSAAATLRVFNANMIKYLRGASS 108
S L S PN++L+ H + ++ + V A + KY+ +S
Sbjct 318 AKNGEGSFNDELNSWTSELVSLPNIILTPHIGGSTEEAQSSIGIEVATA-LSKYINEGNS 376
Query 109 NADMETP 115
+ P
Sbjct 377 VGSVNFP 383
> YIL074c
Length=469
Score = 63.2 bits (152), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 43/127 (33%), Positives = 66/127 (51%), Gaps = 13/127 (10%)
Query 1 LPATAETKQCISYSQFNAMKKNAVFISIGRGSVVDEEALLHALHSGQIAGAALDVFNKEP 60
+PAT ET++ +S QF AMK A I+ RG+VVD +L+ A+ + +IAGAALDV+ EP
Sbjct 258 VPATPETEKMLSAPQFAAMKDGAYVINASRGTVVDIPSLIQAVKANKIAGAALDVYPHEP 317
Query 61 LP------------VDSPLWSSPNLLLSSHNADFCDNFSAAATLRVFNANMIKYLRGASS 108
S L S PN++L+ H + ++ + V A + KY+ +S
Sbjct 318 AKNGEGSFNDELNSWTSELVSLPNIILTPHIGGSTEEAQSSIGIEVATA-LSKYINEGNS 376
Query 109 NADMETP 115
+ P
Sbjct 377 VGSVNFP 383
> At3g19480
Length=588
Score = 62.4 bits (150), Expect = 2e-10, Method: Composition-based stats.
Identities = 41/116 (35%), Positives = 59/116 (50%), Gaps = 1/116 (0%)
Query 1 LPATAETKQCISYSQFNAMKKNAVFISIGRGSVVDEEALLHALHSGQIAGAALDVFNKEP 60
LP TA T + ++ F MKK +++ RG V+DEEALL AL SG +A AALDVF EP
Sbjct 247 LPLTAATSKMMNDVTFAMMKKGVRIVNVARGGVIDEEALLRALDSGIVAQAALDVFTVEP 306
Query 61 LPVDSPLWSSPNLLLSSHNADFCDNFSAAATLRVFNANMIKYLRGASSNADMETPV 116
D+ L ++ + H ++ V A +I LRG + + P+
Sbjct 307 PVKDNKLVLHESVTATPHLGASTMEAQEGVSIEVAEA-VIGALRGELAATAVNAPM 361
> At4g34200
Length=603
Score = 61.6 bits (148), Expect = 4e-10, Method: Composition-based stats.
Identities = 31/78 (39%), Positives = 46/78 (58%), Gaps = 0/78 (0%)
Query 1 LPATAETKQCISYSQFNAMKKNAVFISIGRGSVVDEEALLHALHSGQIAGAALDVFNKEP 60
+P T T + ++ F MKK +++ RG V+DE+AL+ AL +G +A AALDVF KEP
Sbjct 262 MPLTPTTSKILNDETFAKMKKGVRIVNVARGGVIDEDALVRALDAGIVAQAALDVFTKEP 321
Query 61 LPVDSPLWSSPNLLLSSH 78
DS L + ++ H
Sbjct 322 PAKDSKLVQHERVTVTPH 339
> SPCC4G3.01
Length=466
Score = 61.2 bits (147), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 31/60 (51%), Positives = 42/60 (70%), Gaps = 0/60 (0%)
Query 1 LPATAETKQCISYSQFNAMKKNAVFISIGRGSVVDEEALLHALHSGQIAGAALDVFNKEP 60
+PA+ ETK IS +F AMK+ + I+ RG+VVD AL+ A SG+IAGAA+DV+ EP
Sbjct 255 VPASPETKNMISSKEFAAMKEGSYLINASRGTVVDIPALVDASKSGKIAGAAIDVYPSEP 314
> At1g79870
Length=313
Score = 61.2 bits (147), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 31/77 (40%), Positives = 45/77 (58%), Gaps = 1/77 (1%)
Query 2 PATAETKQCISYSQFNAMKKNAVFISIGRGSVVDEEALLHALHSGQIAGAALDVFNKEPL 61
P T +T+ + +A+ V I+IGRG VDE+ L+ AL G++ GAALDVF +EP
Sbjct 204 PLTEQTRHIVDRQVMDALGAKGVLINIGRGPHVDEQELIKALTEGRLGGAALDVFEQEP- 262
Query 62 PVDSPLWSSPNLLLSSH 78
V L+ N++L H
Sbjct 263 HVPEELFGLENVVLLPH 279
> At2g45630
Length=327
Score = 58.5 bits (140), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 34/111 (30%), Positives = 56/111 (50%), Gaps = 6/111 (5%)
Query 6 ETKQCISYSQFNAMKKNAVFISIGRGSVVDEEALLHALHSGQIAGAALDVFNKEPLPVDS 65
+T + I+ +A+ K V +++ RG+++DEE ++ L G+I GA LDVF EP V
Sbjct 222 KTLRLINKDVLSALGKRGVIVNVARGAIIDEEEMVRCLREGEIGGAGLDVFEDEP-NVPK 280
Query 66 PLWSSPNLLLSSHNADFCDNFSAAATLRVFNANMIKYLRGASSNADMETPV 116
L+ N++ S H+A F L ++ + SN + TPV
Sbjct 281 ELFELDNVVFSPHSA-----FMTLEGLEELGKVVVGNIEAFFSNKPLLTPV 326
> Hs4557499
Length=445
Score = 58.2 bits (139), Expect = 4e-09, Method: Composition-based stats.
Identities = 33/79 (41%), Positives = 45/79 (56%), Gaps = 6/79 (7%)
Query 19 MKKNAVFISIGRGSVVDEEALLHALHSGQIAGAALDVFNKEPLP-VDSPLWSSPNLLLSS 77
M++ A ++ RG +VDE+AL AL G+I GAALDV EP PL +PNL+ +
Sbjct 261 MRQGAFLVNAARGGLVDEKALAQALKEGRIRGAALDVHESEPFSFAQGPLKDAPNLICTP 320
Query 78 HNADFCDNFS-----AAAT 91
H A + + S AAAT
Sbjct 321 HTAWYSEQASLEMREAAAT 339
> Hs5729974
Length=533
Score = 57.8 bits (138), Expect = 5e-09, Method: Composition-based stats.
Identities = 33/77 (42%), Positives = 42/77 (54%), Gaps = 1/77 (1%)
Query 2 PATAETKQCISYSQFNAMKKNAVFISIGRGSVVDEEALLHALHSGQIAGAALDVFNKEPL 61
P T ++ + F KK ++ RG +VDE ALL AL SGQ AGAALDVF +EP
Sbjct 208 PLLPSTTGLLNDNTFAQCKKGVRVVNCARGGIVDEGALLRALQSGQCAGAALDVFTEEP- 266
Query 62 PVDSPLWSSPNLLLSSH 78
P D L N++ H
Sbjct 267 PRDRALVDHENVISCPH 283
> Hs14744596
Length=533
Score = 57.8 bits (138), Expect = 5e-09, Method: Composition-based stats.
Identities = 33/77 (42%), Positives = 42/77 (54%), Gaps = 1/77 (1%)
Query 2 PATAETKQCISYSQFNAMKKNAVFISIGRGSVVDEEALLHALHSGQIAGAALDVFNKEPL 61
P T ++ + F KK ++ RG +VDE ALL AL SGQ AGAALDVF +EP
Sbjct 208 PLLPSTTGLLNDNTFAQCKKGVRVVNCARGGIVDEGALLRALQSGQCAGAALDVFTEEP- 266
Query 62 PVDSPLWSSPNLLLSSH 78
P D L N++ H
Sbjct 267 PRDRALVDHENVISCPH 283
> SPBC1773.17c
Length=185
Score = 57.8 bits (138), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 32/102 (31%), Positives = 51/102 (50%), Gaps = 2/102 (1%)
Query 2 PATAETKQCISYSQFNAMKKNAVFISIGRGSVVDEEALLHALHSGQIAGAALDVFNKEPL 61
P T T IS +F MK I+ RG++++E+A + A+ SG++A A LDVF EP
Sbjct 68 PLTPATHDLISTKEFEKMKDGVYIINTARGAIINEDAFIKAIKSGKVARAGLDVFLNEPT 127
Query 62 PVDSPLWSSPNLLLSSHNADFCDNFSAAATLRVFNANMIKYL 103
P + W + + + NF+ A T A++ +L
Sbjct 128 P--NKFWLECDKVTIQPHCGVYTNFTVAKTEECVLASIETFL 167
> SPACUNK4.10
Length=334
Score = 57.0 bits (136), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 29/78 (37%), Positives = 43/78 (55%), Gaps = 1/78 (1%)
Query 1 LPATAETKQCISYSQFNAMKKNAVFISIGRGSVVDEEALLHALHSGQIAGAALDVFNKEP 60
LP A T+ I +F MK+ V ++ RG+V+DE AL+ AL G + A LDVF +EP
Sbjct 217 LPLNAHTRHIIGKPEFQKMKRGIVIVNTARGAVMDEAALVEALDEGIVYSAGLDVFEEEP 276
Query 61 LPVDSPLWSSPNLLLSSH 78
+ L + ++L H
Sbjct 277 -KIHPGLLENEKVILLPH 293
> Hs4557497
Length=440
Score = 56.6 bits (135), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 29/70 (41%), Positives = 41/70 (58%), Gaps = 1/70 (1%)
Query 19 MKKNAVFISIGRGSVVDEEALLHALHSGQIAGAALDVFNKEPLPV-DSPLWSSPNLLLSS 77
M++ A ++ RG +VDE+AL AL G+I GAALDV EP PL +PNL+ +
Sbjct 255 MRQGAFLVNTARGGLVDEKALAQALKEGRIRGAALDVHESEPFSFSQGPLKDAPNLICTP 314
Query 78 HNADFCDNFS 87
H A + + S
Sbjct 315 HAAWYSEQAS 324
> At1g12550
Length=323
Score = 55.5 bits (132), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 29/77 (37%), Positives = 43/77 (55%), Gaps = 1/77 (1%)
Query 4 TAETKQCISYSQFNAMKKNAVFISIGRGSVVDEEALLHALHSGQIAGAALDVFNKEPLPV 63
T ET ++ + K+ V I++GRG ++DE+ ++ L G I GA LDVF EP V
Sbjct 214 TDETHHIVNREVMELLGKDGVVINVGRGKLIDEKEMVKCLVDGVIGGAGLDVFENEP-AV 272
Query 64 DSPLWSSPNLLLSSHNA 80
L+ N++LS H A
Sbjct 273 PQELFGLDNVVLSPHFA 289
> Hs12746590
Length=985
Score = 55.5 bits (132), Expect = 2e-08, Method: Composition-based stats.
Identities = 33/79 (41%), Positives = 45/79 (56%), Gaps = 6/79 (7%)
Query 19 MKKNAVFISIGRGSVVDEEALLHALHSGQIAGAALDVFNKEPLP-VDSPLWSSPNLLLSS 77
M++ A ++ RG +VDE+AL AL G+I GAALDV EP PL +PNL+ +
Sbjct 801 MRQGAFLVNAARGGLVDEKALAQALKEGRIRGAALDVHESEPFSFAQGPLKDAPNLICTP 860
Query 78 HNADFCDNFS-----AAAT 91
H A + + S AAAT
Sbjct 861 HTAWYSEQASLEMREAAAT 879
> 7298717
Length=248
Score = 55.5 bits (132), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 27/77 (35%), Positives = 45/77 (58%), Gaps = 0/77 (0%)
Query 2 PATAETKQCISYSQFNAMKKNAVFISIGRGSVVDEEALLHALHSGQIAGAALDVFNKEPL 61
P T +T+ + + FN MK+ AV ++I RG +V+++ L AL + +I A LDV + EPL
Sbjct 138 PLTKDTQGVFNATAFNKMKQTAVLVNIARGKIVNQDDLYEALKANRIFSAGLDVTDPEPL 197
Query 62 PVDSPLWSSPNLLLSSH 78
L + N+++ H
Sbjct 198 SPKDKLLTLDNVVVLPH 214
> 7297831
Length=332
Score = 53.9 bits (128), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 27/79 (34%), Positives = 43/79 (54%), Gaps = 2/79 (2%)
Query 2 PATAETKQCISYSQFNAMKKNAVFISIGRGSVVDEEALLHALHSGQIAGAALDVFNKEPL 61
P T+ IS K+ +++ RG ++DE+A+L L SG++AGAA DV+ +EP
Sbjct 208 PLIPATRNLISAETLAKCKQGVKVVNVARGGIIDEQAVLDGLESGKVAGAAFDVYPEEPP 267
Query 62 P--VDSPLWSSPNLLLSSH 78
V L S P ++ + H
Sbjct 268 KSAVTKALISHPKVVATPH 286
> YPL113c
Length=396
Score = 53.1 bits (126), Expect = 1e-07, Method: Composition-based stats.
Identities = 26/67 (38%), Positives = 36/67 (53%), Gaps = 0/67 (0%)
Query 1 LPATAETKQCISYSQFNAMKKNAVFISIGRGSVVDEEALLHALHSGQIAGAALDVFNKEP 60
LP+TA T I+ K +++GRG+ +DE+ LL AL SG++A LDVF E
Sbjct 284 LPSTASTNNIINRKSLAWCKDGVRIVNVGRGTCIDEDVLLDALESGKVASCGLDVFKNEE 343
Query 61 LPVDSPL 67
V L
Sbjct 344 TRVKQEL 350
> SPAC186.07c
Length=332
Score = 52.8 bits (125), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 31/94 (32%), Positives = 49/94 (52%), Gaps = 13/94 (13%)
Query 2 PATAETKQCISYSQFNAMKKNAVFISIGRGSVVDEEALLHALHSGQIAGAALDVFNKE-- 59
P T +T+ + +MKK I+ RG +VD +AL+ A+ SGQ+ G A+DV+ E
Sbjct 207 PLTPDTEHLVDEKLLASMKKGVKIINTSRGGLVDTKALVKAIESGQVGGCAMDVYEGERR 266
Query 60 --------PLPVDSP---LWSSPNLLLSSHNADF 82
+ D+ L + PN+L++SH A F
Sbjct 267 LFYRDLSNEVIKDTTFQQLANFPNVLVTSHQAFF 300
> 7299709
Length=386
Score = 52.8 bits (125), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 33/84 (39%), Positives = 45/84 (53%), Gaps = 3/84 (3%)
Query 19 MKKNAVFISIGRGSVVDEEALLHALHSGQIAGAALDVFNKEPLPV-DSPLWSSPNLLLSS 77
M+ A ++ RG +VD+E L AL G+I AALDV EP V L +PNL+ +
Sbjct 255 MRPGAFLVNTARGGLVDDETLALALKQGRIRAAALDVHENEPYNVFQGALKDAPNLICTP 314
Query 78 HNADFCDNFSAAATLRVFNANMIK 101
H A F D ++A LR A I+
Sbjct 315 HAAFFSD--ASATELREMAATEIR 336
> At5g14780
Length=384
Score = 52.4 bits (124), Expect = 2e-07, Method: Composition-based stats.
Identities = 22/80 (27%), Positives = 45/80 (56%), Gaps = 0/80 (0%)
Query 1 LPATAETKQCISYSQFNAMKKNAVFISIGRGSVVDEEALLHALHSGQIAGAALDVFNKEP 60
+P T +T+ + +KK + ++ RG++++ +A++ A+ SG I G + DV++ +P
Sbjct 261 MPLTEKTRGMFNKELIGKLKKGVLIVNNARGAIMERQAVVDAVESGHIGGYSGDVWDPQP 320
Query 61 LPVDSPLWSSPNLLLSSHNA 80
P D P PN ++ H +
Sbjct 321 APKDHPWRYMPNQAMTPHTS 340
> SPAC186.02c
Length=332
Score = 51.2 bits (121), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 30/94 (31%), Positives = 47/94 (50%), Gaps = 13/94 (13%)
Query 2 PATAETKQCISYSQFNAMKKNAVFISIGRGSVVDEEALLHALHSGQIAGAALDVFNKE-- 59
P T T ++ MKK ++ RG ++D +AL+ A+ SGQ+ G A+DV+ E
Sbjct 207 PLTPSTTHIVNSDSLALMKKGVTIVNTSRGGLIDTKALVDAIDSGQVGGCAIDVYEGERN 266
Query 60 --------PLPVDSP---LWSSPNLLLSSHNADF 82
+ DS L + PN+L++SH A F
Sbjct 267 LFYKDLSNEVIKDSTFQRLVNFPNVLVTSHQAFF 300
> YNL274c
Length=350
Score = 51.2 bits (121), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 24/60 (40%), Positives = 33/60 (55%), Gaps = 0/60 (0%)
Query 1 LPATAETKQCISYSQFNAMKKNAVFISIGRGSVVDEEALLHALHSGQIAGAALDVFNKEP 60
+P T I+ MK V ++ RG+V+DE+A+ AL SG+I A LDVF EP
Sbjct 225 VPLNHNTHHLINAETIEKMKDGVVIVNTARGAVIDEQAMTDALRSGKIRSAGLDVFEYEP 284
> CE08497
Length=322
Score = 49.7 bits (117), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 34/109 (31%), Positives = 55/109 (50%), Gaps = 10/109 (9%)
Query 1 LPATAETKQCISYSQFNAMKKNAVFISIGRGSVVDEEALLHALHSGQIAGAALDVFNKEP 60
+P +T+ I+ KK I++ RG +V+E L+ +L++G GAA DVF +EP
Sbjct 207 VPLIKQTENLINKETLAKCKKGVRIINVARGGIVNEVDLVESLNAGHAKGAAFDVFEQEP 266
Query 61 LPVDSPLWSSPNLLLSSH-NADFCDNFSAAATLRVFNA---NMIKYLRG 105
P L P ++ + H A D A LRV + N+++Y +G
Sbjct 267 -PTFRELIDHPLVIATPHLGASTID-----AQLRVASEIADNIVQYNKG 309
> At1g72190
Length=344
Score = 48.5 bits (114), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 23/80 (28%), Positives = 42/80 (52%), Gaps = 0/80 (0%)
Query 1 LPATAETKQCISYSQFNAMKKNAVFISIGRGSVVDEEALLHALHSGQIAGAALDVFNKEP 60
L ET + ++ +MKK A+ ++I RG +++ E+ L SG + G +DV EP
Sbjct 231 LRLNKETAEIVNKEFICSMKKGALLVNIARGGLINYESAFQNLESGHLGGLGIDVAWSEP 290
Query 61 LPVDSPLWSSPNLLLSSHNA 80
+ P+ N++++ H A
Sbjct 291 FDPNDPILKFKNVIITPHVA 310
> YGL185c
Length=379
Score = 48.1 bits (113), Expect = 4e-06, Method: Composition-based stats.
Identities = 26/73 (35%), Positives = 40/73 (54%), Gaps = 1/73 (1%)
Query 1 LPATAETKQCISYSQFNAMKKNAVFISIGRGSVVDEEALLHALHSGQIAGAALDVFNKEP 60
LP T +T+ I+ + +++GRG ++D A+ AL +G+I LDVFNKEP
Sbjct 264 LPGTPQTEHLINRKFLEHCNPGLILVNLGRGKILDLRAVSDALVTGRINHLGLDVFNKEP 323
Query 61 LPVDSPLWSSPNL 73
+D + SS L
Sbjct 324 -EIDEKIRSSDRL 335
> Hs20545061
Length=200
Score = 47.0 bits (110), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 23/63 (36%), Positives = 35/63 (55%), Gaps = 1/63 (1%)
Query 19 MKKNAVFISIGRGSVVDEEALLHALHSGQIAGAALDVFNKEPLP-VDSPLWSSPNLLLSS 77
M++ A ++ G +VDE+AL L +I GAAL + EP +PL +PNL+ +
Sbjct 138 MRQGAFLVNAAHGGLVDEKALAQVLKEDRIGGAALGLHESEPFSFAQTPLKDAPNLIFTP 197
Query 78 HNA 80
H A
Sbjct 198 HTA 200
> YPL275w
Length=236
Score = 42.4 bits (98), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 20/69 (28%), Positives = 37/69 (53%), Gaps = 1/69 (1%)
Query 2 PATAETKQCISYSQFNAMKKNAVFISIGRGSVVDEEALLHALHSGQIAGAALDVFNKEPL 61
P +++ + + MK A ++ RG++ E + A+ SG++AG DV++K+P
Sbjct 104 PLHKDSRGLFNKKLISHMKDGAYLVNTARGAICVAEDVAEAVKSGKLAGYGGDVWDKQPA 163
Query 62 PVDSPLWSS 70
P D P W +
Sbjct 164 PKDHP-WRT 171
> YOR388c
Length=376
Score = 42.0 bits (97), Expect = 3e-04, Method: Composition-based stats.
Identities = 20/69 (28%), Positives = 37/69 (53%), Gaps = 1/69 (1%)
Query 2 PATAETKQCISYSQFNAMKKNAVFISIGRGSVVDEEALLHALHSGQIAGAALDVFNKEPL 61
P +++ + + MK A ++ RG++ E + A+ SG++AG DV++K+P
Sbjct 244 PLHKDSRGLFNKKLISHMKDGAYLVNTARGAICVAEDVAEAVKSGKLAGYGGDVWDKQPA 303
Query 62 PVDSPLWSS 70
P D P W +
Sbjct 304 PKDHP-WRT 311
> At1g68010
Length=386
Score = 41.2 bits (95), Expect = 4e-04, Method: Composition-based stats.
Identities = 19/55 (34%), Positives = 29/55 (52%), Gaps = 0/55 (0%)
Query 7 TKQCISYSQFNAMKKNAVFISIGRGSVVDEEALLHALHSGQIAGAALDVFNKEPL 61
T ++ + MKK A+ ++ RG V+DE AL+ L + LDVF +EP
Sbjct 250 TYHLVNKERLAMMKKEAILVNCSRGPVIDEAALVEHLKENPMFRVGLDVFEEEPF 304
> CE28436
Length=701
Score = 36.6 bits (83), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 26/103 (25%), Positives = 46/103 (44%), Gaps = 7/103 (6%)
Query 6 ETKQCISYSQFNAMKKNAVFISIGRGSVVDEEALLHALHSGQIAGAALDVFNKEPLPVD- 64
ET+ I+ K ++ +++E L AL +G + GAALDV + +
Sbjct 367 ETRGIINADSLRQCKSGVYIVNTSHAGLINENDLAAALKNGHVKGAALDVHDSVRFDPNC 426
Query 65 -SPLWSSPNLLLSSHNADFCDNFSAAATLRVFNANMIKYLRGA 106
+PL PN++ + H+A + A+ + N K +R A
Sbjct 427 LNPLVGCPNIINTPHSA-----WMTEASCKDLRINAAKEIRKA 464
> At1g01510
Length=634
Score = 34.3 bits (77), Expect = 0.051, Method: Compositional matrix adjust.
Identities = 23/83 (27%), Positives = 38/83 (45%), Gaps = 3/83 (3%)
Query 4 TAETKQCISYSQFNAMKKNAVFISIGRGSVVDEEALLHALHSGQIAGAALDVFNKEPLPV 63
T +T Q ++ +K A ++ G ++D+ A+ L G IAG ALD P
Sbjct 234 TNDTVQILNAECLQHIKPGAFLVNTGSCQLLDDCAVKQLLIDGTIAGCALDGAEG---PQ 290
Query 64 DSPLWSSPNLLLSSHNADFCDNF 86
W PN+L+ +AD+ +
Sbjct 291 WMEAWEMPNVLILPRSADYSEEV 313
> Hs20558290
Length=515
Score = 33.1 bits (74), Expect = 0.12, Method: Composition-based stats.
Identities = 14/32 (43%), Positives = 23/32 (71%), Gaps = 0/32 (0%)
Query 19 MKKNAVFISIGRGSVVDEEALLHALHSGQIAG 50
M + A ++ RG++VDE+AL AL++G+I G
Sbjct 311 MSQGAFLVNAARGALVDEKALAQALNNGKIRG 342
> Hs22060587
Length=356
Score = 29.3 bits (64), Expect = 1.9, Method: Composition-based stats.
Identities = 14/32 (43%), Positives = 18/32 (56%), Gaps = 0/32 (0%)
Query 30 RGSVVDEEALLHALHSGQIAGAALDVFNKEPL 61
+G+ + A H L G+I GAALDV EP
Sbjct 139 QGAFLVNAACAHTLKEGRIRGAALDVNESEPF 170
> 7297700
Length=674
Score = 28.9 bits (63), Expect = 2.5, Method: Composition-based stats.
Identities = 10/22 (45%), Positives = 15/22 (68%), Gaps = 0/22 (0%)
Query 64 DSPLWSSPNLLLSSHNADFCDN 85
D+P W++P LL HN +F D+
Sbjct 221 DNPKWTTPELLAFKHNLEFPDS 242
> At1g33410
Length=1459
Score = 27.3 bits (59), Expect = 6.4, Method: Compositional matrix adjust.
Identities = 10/20 (50%), Positives = 15/20 (75%), Gaps = 0/20 (0%)
Query 62 PVDSPLWSSPNLLLSSHNAD 81
P LWSSPNL+L+S++ +
Sbjct 1258 PAKGTLWSSPNLMLTSNDEE 1277
Lambda K H
0.316 0.129 0.372
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1171470346
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40