bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_1220_orf2
Length=164
Score E
Sequences producing significant alignments: (Bits) Value
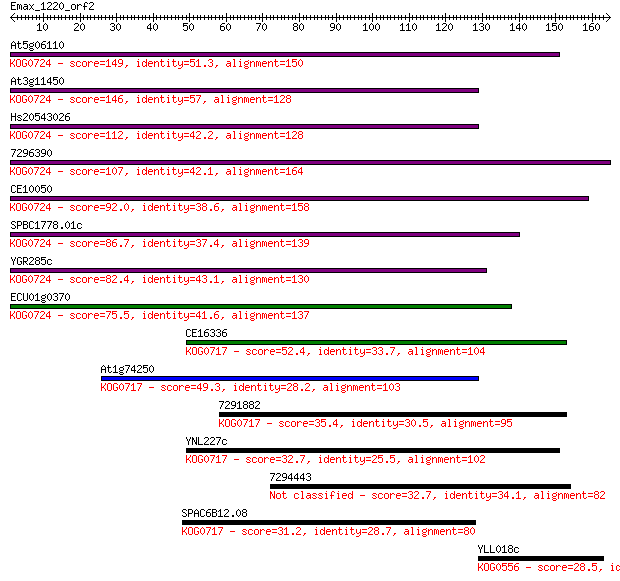
At5g06110 149 2e-36
At3g11450 146 2e-35
Hs20543026 112 3e-25
7296390 107 1e-23
CE10050 92.0 5e-19
SPBC1778.01c 86.7 2e-17
YGR285c 82.4 4e-16
ECU01g0370 75.5 4e-14
CE16336 52.4 4e-07
At1g74250 49.3 3e-06
7291882 35.4 0.050
YNL227c 32.7 0.32
7294443 32.7 0.33
SPAC6B12.08 31.2 1.00
YLL018c 28.5 5.7
> At5g06110
Length=663
Score = 149 bits (377), Expect = 2e-36, Method: Compositional matrix adjust.
Identities = 77/150 (51%), Positives = 109/150 (72%), Gaps = 4/150 (2%)
Query 1 RRQYDSALPFDESVPSAADCQSPEDFYKIFGAAFQRNARWSVNRPVLTLGDCSTPLSAVE 60
RR +DS FD+ VP+ DC +P+DF+K+FG AF+RNARWS N P+ LGD +TPL V+
Sbjct 175 RRIFDSTDEFDDKVPT--DC-APQDFFKVFGPAFKRNARWS-NSPLPDLGDENTPLKEVD 230
Query 61 EFYEFWFAFESWRDFGVHDEHDLDQAACRLERRWMERENARIKKKYIKNERARIQKLVET 120
FY W+ F+SWR+F +EHD++QA R E+RWMERENAR +K K E ARI+ LV+
Sbjct 231 RFYSTWYTFKSWREFPEEEEHDIEQAESREEKRWMERENARKTQKARKEEYARIRTLVDN 290
Query 121 AHAADPRIKQRKEEERKKKEEEKAAQLRAR 150
A+ D RI++RK++E+ KK ++K A++ A+
Sbjct 291 AYKKDIRIQKRKDDEKAKKLQKKEAKVMAK 320
> At3g11450
Length=663
Score = 146 bits (369), Expect = 2e-35, Method: Compositional matrix adjust.
Identities = 73/128 (57%), Positives = 94/128 (73%), Gaps = 3/128 (2%)
Query 1 RRQYDSALPFDESVPSAADCQSPEDFYKIFGAAFQRNARWSVNRPVLTLGDCSTPLSAVE 60
RR +DS FD+ VPS DC P+DF+K+FG AF+RNARWSVN+ + LGD +TPL V+
Sbjct 190 RRIFDSTDEFDDEVPS--DCL-PQDFFKVFGPAFKRNARWSVNQRIPDLGDENTPLKDVD 246
Query 61 EFYEFWFAFESWRDFGVHDEHDLDQAACRLERRWMERENARIKKKYIKNERARIQKLVET 120
+FY FW+AF+SWR+F +EHDL+QA R ERRWME+ENA+ K K E ARI+ LV+
Sbjct 247 KFYNFWYAFKSWREFPDEEEHDLEQADSREERRWMEKENAKKTVKARKEEHARIRTLVDN 306
Query 121 AHAADPRI 128
A+ DPRI
Sbjct 307 AYRKDPRI 314
> Hs20543026
Length=621
Score = 112 bits (280), Expect = 3e-25, Method: Compositional matrix adjust.
Identities = 54/129 (41%), Positives = 85/129 (65%), Gaps = 3/129 (2%)
Query 1 RRQYDSALP-FDESVPSAADCQSPEDFYKIFGAAFQRNARWSVNRPVLTLGDCSTPLSAV 59
RR ++S P FD SVPS ++ + ++F+++F F+RN+RWS + V LGD ++ V
Sbjct 154 RRAFNSVDPTFDNSVPSKSEAK--DNFFEVFTPVFERNSRWSNKKNVPKLGDMNSSFEDV 211
Query 60 EEFYEFWFAFESWRDFGVHDEHDLDQAACRLERRWMERENARIKKKYIKNERARIQKLVE 119
+ FY FW+ F+SWR+F DE + ++A CR ERRW+E++N + + K E RI+ LV+
Sbjct 212 DIFYSFWYNFDSWREFSYLDEEEKEKAECRDERRWIEKQNRATRAQRKKEEMNRIRTLVD 271
Query 120 TAHAADPRI 128
A++ DPRI
Sbjct 272 NAYSCDPRI 280
> 7296390
Length=646
Score = 107 bits (267), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 69/167 (41%), Positives = 100/167 (59%), Gaps = 8/167 (4%)
Query 1 RRQYDSALP-FDESVPSAADCQSPEDFYKIFGAAFQRNARWSVNRPVLTLGDCSTPLSAV 59
RR +DS P FD+S+PS D + D++ +F F N RWS V + G V
Sbjct 141 RRSFDSVDPEFDDSLPSQNDIDN--DYFGVFNKFFTLNGRWSEKPHVPSFGQVDAKREEV 198
Query 60 EEFYEFWFAFESWRDFGVHDEHDLDQAACRLERRWMEREN--ARIKKKYIKNERARIQKL 117
E FY FW+ F+SWR+F DE D ++ R ERRW+E+EN ARIK+K K E +RI+ L
Sbjct 199 ERFYNFWYDFKSWREFSYLDEEDKEKGQDRDERRWIEKENRAARIKRK--KEEMSRIRSL 256
Query 118 VETAHAADPRIKQRKEEERKKKEEEKAAQLRAREEARRAAEDQKRRE 164
V+ A+ D RI++ K+EE+ +K K A++ A +A++A D+ RE
Sbjct 257 VDLAYNNDKRIQRFKQEEKDRKAAAKRAKMDA-AQAQKAEADRAIRE 302
> CE10050
Length=589
Score = 92.0 bits (227), Expect = 5e-19, Method: Compositional matrix adjust.
Identities = 61/159 (38%), Positives = 92/159 (57%), Gaps = 2/159 (1%)
Query 1 RRQYDSA-LPFDESVPSAADCQSPEDFYKIFGAAFQRNARWSVNRPVLTLGDCSTPLSAV 59
R+ +DS F++ +P+ +FY FQ N+RWS +PV LG V
Sbjct 163 RQAFDSVDHKFNDIIPNEKSINH-NNFYNELAPVFQLNSRWSNIKPVPELGKSDATREDV 221
Query 60 EEFYEFWFAFESWRDFGVHDEHDLDQAACRLERRWMERENARIKKKYIKNERARIQKLVE 119
E FY+FWF F+SWR+F DE D ++ R ERR ME++N +++ K E RI+KLV+
Sbjct 222 ENFYDFWFNFQSWREFSYLDEEDKERGEDRYERREMEKQNKAERERRRKEEAKRIRKLVD 281
Query 120 TAHAADPRIKQRKEEERKKKEEEKAAQLRAREEARRAAE 158
A+A DPRI + K+E++ KK++ K + RA E + A +
Sbjct 282 IAYAKDPRIIKFKKEQQAKKDKAKEDKQRAIREKQEAID 320
> SPBC1778.01c
Length=354
Score = 86.7 bits (213), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 52/144 (36%), Positives = 81/144 (56%), Gaps = 12/144 (8%)
Query 1 RRQYDSALPFDESVPSAADCQSPED-----FYKIFGAAFQRNARWSVNRPVLTLGDCSTP 55
RRQ+DS V AD + PE F++++ F+ AR+S +PV +LG +
Sbjct 72 RRQFDS-------VDENADVEPPESTTKETFFELWTPVFESEARFSKKQPVPSLGTIEST 124
Query 56 LSAVEEFYEFWFAFESWRDFGVHDEHDLDQAACRLERRWMERENARIKKKYIKNERARIQ 115
+ V+ FY FW+ F+SWR F D+ D R +R+ E++N ++K + AR++
Sbjct 125 RAEVDNFYNFWYNFDSWRSFEYLDKDIPDDGESRDNKRFQEKKNRSERQKNKARDNARLR 184
Query 116 KLVETAHAADPRIKQRKEEERKKK 139
LV+TA A+DPRIK KE+E+ K
Sbjct 185 NLVDTALASDPRIKLFKEQEKAAK 208
> YGR285c
Length=433
Score = 82.4 bits (202), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 56/132 (42%), Positives = 75/132 (56%), Gaps = 6/132 (4%)
Query 1 RRQYDSALPFDESVPSAADCQSPEDFYKIFGAAFQRNARWSVNRPVLTLGDCSTPLSAVE 60
R QYDS F VP + DFY+ +G F+ AR+S P+ +LG+ + VE
Sbjct 161 RAQYDSC-DFVADVPPPKKG-TDYDFYEAWGPVFEAEARFSKKTPIPSLGNKDSSKKEVE 218
Query 61 EFYEFWFAFESWRDFGVHDEHDLDQAACRLERRWMEREN--ARIKKKYIKNERARIQKLV 118
+FY FW F+SWR F DE D ++ R +R++ER+N AR KKK N AR+ KLV
Sbjct 219 QFYAFWHRFDSWRTFEFLDEDVPDDSSNRDHKRYIERKNKAARDKKKTADN--ARLVKLV 276
Query 119 ETAHAADPRIKQ 130
E A + DPRIK
Sbjct 277 ERAVSEDPRIKM 288
> ECU01g0370
Length=295
Score = 75.5 bits (184), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 57/145 (39%), Positives = 79/145 (54%), Gaps = 22/145 (15%)
Query 1 RRQYDSALPFDESVPSAADCQSPEDFYKIFGAAFQRNARWSVNRPVLTLGDCSTPLSAVE 60
R YDS FDES+P Q P++F+ +F F+RN+++S+ +PV L S L VE
Sbjct 104 RLFYDSNF-FDESIPEDRIYQ-PDEFFDVFEECFRRNSKFSIKQPVPLLSP-SDDLKKVE 160
Query 61 EFYEFWFAFESWRDF-------GVHDEHDLDQAACRLERRWMERENARIKKKYIKNERA- 112
EFYEFW F SWR F G+ +EHD Q + + R K +KN+ A
Sbjct 161 EFYEFWSNFRSWRTFEPVEELYGM-EEHDRSQYSAK----------NREKLASLKNQDAL 209
Query 113 RIQKLVETAHAADPRIKQRKEEERK 137
RI++LV+ A DPRI + EE+ K
Sbjct 210 RIKRLVQIAKKRDPRIGKSIEEQIK 234
> CE16336
Length=510
Score = 52.4 bits (124), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 35/112 (31%), Positives = 55/112 (49%), Gaps = 8/112 (7%)
Query 49 LGDCSTPL-SAVEEFYEFWFAFESWRDFGVHDEHDLDQAACRLERRWMERENARIKKKYI 107
GD T L V FY FW +F + R F D +D+ QA+ R E R +++EN + +
Sbjct 160 FGDKDTDLEQTVNGFYGFWSSFSTTRSFAWLDHYDITQASNRFESRQIDQENKKFRDVGK 219
Query 108 KNERARIQKLVETAHAADPRIK-------QRKEEERKKKEEEKAAQLRAREE 152
+ +I+ LV DPR+K Q+K E KK+ + + Q+ +E
Sbjct 220 QERNEQIRNLVAFVRKRDPRVKAYREILEQKKLEAHKKQADNRKKQIAKNQE 271
> At1g74250
Length=630
Score = 49.3 bits (116), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 29/111 (26%), Positives = 47/111 (42%), Gaps = 8/111 (7%)
Query 26 FYKIFGAAFQR--------NARWSVNRPVLTLGDCSTPLSAVEEFYEFWFAFESWRDFGV 77
+Y +F + + R R +G+ +P + V FY +W F + DF
Sbjct 127 YYDVFNSVYLNEIKFARTLGLRMDSVREAPIMGNLESPYAQVTAFYNYWLGFCTVMDFCW 186
Query 78 HDEHDLDQAACRLERRWMERENARIKKKYIKNERARIQKLVETAHAADPRI 128
DE+D+ R RR ME EN + +KK + ++ L E D R+
Sbjct 187 VDEYDVMGGPNRKSRRMMEEENKKSRKKAKREYNDTVRGLAEFVKKRDKRV 237
> 7291882
Length=540
Score = 35.4 bits (80), Expect = 0.050, Method: Compositional matrix adjust.
Identities = 29/102 (28%), Positives = 49/102 (48%), Gaps = 7/102 (6%)
Query 58 AVEEFYEFWFAFESWRDFGVHDEHDLDQAACRLERRWMERENARIKKKYIKNERARIQKL 117
V FY FW A+ + + + +D+ + R R +E+E +I + K ++ L
Sbjct 147 VVGPFYAFWQAYSTRKTYDWLCPYDVREIKERFILRKVEKEMKKIVQAARKERNEEVRNL 206
Query 118 VETAHAADPRIK-------QRKEEERKKKEEEKAAQLRAREE 152
V DPR++ +R E R K+EE++ QLR R+E
Sbjct 207 VNFVRKRDPRVQAYRRMLEERVEANRLKQEEKRKEQLRKRQE 248
> YNL227c
Length=590
Score = 32.7 bits (73), Expect = 0.32, Method: Compositional matrix adjust.
Identities = 26/105 (24%), Positives = 49/105 (46%), Gaps = 3/105 (2%)
Query 49 LGDCSTPLSAVEEFYEFWFAFESWRDFGVHDEHDLDQAACRLERRWMERENARIKKKYIK 108
G T ++ FY+ W AF + + F DE+ + R +R + R N + +++
Sbjct 181 FGYSPTDYEYLKHFYKTWSAFNTLKSFSWKDEYMYSKNYDRRTKREVNRRNEKARQQARN 240
Query 109 NERARIQKLVETAHAADPRIKQR---KEEERKKKEEEKAAQLRAR 150
+++ V D R+K+ EE+RK KE+++ +L R
Sbjct 241 EYNKTVKRFVVFIKKLDKRMKEGAKIAEEQRKLKEQQRKNELNNR 285
> 7294443
Length=1265
Score = 32.7 bits (73), Expect = 0.33, Method: Compositional matrix adjust.
Identities = 28/94 (29%), Positives = 47/94 (50%), Gaps = 12/94 (12%)
Query 72 WRDFGVHDEHDLDQAACRLERR-----WMERENARIKKKY--IKNERARIQKLVETAHAA 124
RD HD+HD+ + + LE + +ER ++ K ++ A +Q+ V+ A A
Sbjct 400 LRDLSAHDKHDIQKLSKELEMKRSEVTELERTKEKLSAKIDELEAIVADLQEQVDAALGA 459
Query 125 DPRIKQRKE-----EERKKKEEEKAAQLRAREEA 153
+ ++Q E E++ K EE+ AQL A EE
Sbjct 460 EEMVEQLAEKKMELEDKVKLLEEEIAQLEALEEV 493
> SPAC6B12.08
Length=380
Score = 31.2 bits (69), Expect = 1.00, Method: Compositional matrix adjust.
Identities = 23/83 (27%), Positives = 40/83 (48%), Gaps = 6/83 (7%)
Query 48 TLGDCSTPLSAVEEFYEFWFAFESWRDF---GVHDEHDLDQAACRLERRWMERENARIKK 104
TLGD + + + Y+ W F + + F +++E + AA R R M+R+N R +
Sbjct 123 TLGDTTWLWTYAKPIYQKWLRFSTKKSFEWEALYNEEEESDAATR---RLMKRQNQRQIQ 179
Query 105 KYIKNERARIQKLVETAHAADPR 127
I+ ++ L+ A DPR
Sbjct 180 YCIQRYNELVRDLIGKACDLDPR 202
> YLL018c
Length=557
Score = 28.5 bits (62), Expect = 5.7, Method: Compositional matrix adjust.
Identities = 18/37 (48%), Positives = 26/37 (70%), Gaps = 3/37 (8%)
Query 129 KQRKEEERKKKEEEKAAQLRAREEARR---AAEDQKR 162
K +KE+E+++K+EE+A QL A EAR AAED +
Sbjct 36 KLQKEQEKQRKKEERALQLEAEREAREKKAAAEDTAK 72
Lambda K H
0.318 0.130 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2317343104
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40