bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_1224_orf1
Length=118
Score E
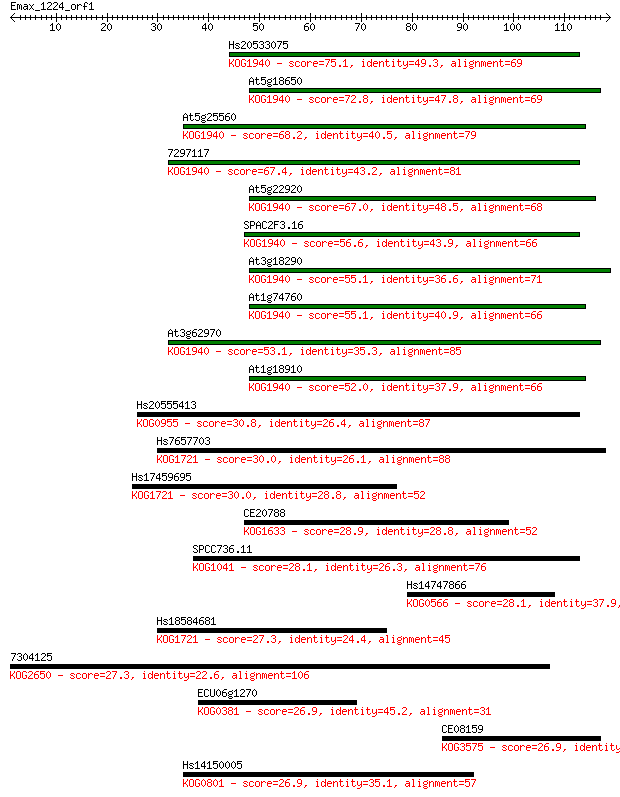
Sequences producing significant alignments: (Bits) Value
Hs20533075 75.1 3e-14
At5g18650 72.8 1e-13
At5g25560 68.2 3e-12
7297117 67.4 6e-12
At5g22920 67.0 9e-12
SPAC2F3.16 56.6 1e-08
At3g18290 55.1 3e-08
At1g74760 55.1 3e-08
At3g62970 53.1 1e-07
At1g18910 52.0 2e-07
Hs20555413 30.8 0.68
Hs7657703 30.0 1.1
Hs17459695 30.0 1.1
CE20788 28.9 2.7
SPCC736.11 28.1 3.6
Hs14747866 28.1 4.3
Hs18584681 27.3 6.6
7304125 27.3 7.1
ECU06g1270 26.9 8.0
CE08159 26.9 8.4
Hs14150005 26.9 10.0
> Hs20533075
Length=261
Score = 75.1 bits (183), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 34/70 (48%), Positives = 45/70 (64%), Gaps = 1/70 (1%)
Query 44 QAIRCPVCSKSIADYSEFWKQLSDEIARTPMEDDMKR-KVRIACNDCLERSTTDFHFLGL 102
+ RCP+C S D + +W+QL DE+A+TPM + + V I CNDC RST FH LG+
Sbjct 179 EGYRCPLCMHSALDMTRYWRQLDDEVAQTPMPSEYQNMTVDILCNDCNGRSTVQFHILGM 238
Query 103 KCMHCNSFNT 112
KC C S+NT
Sbjct 239 KCKICESYNT 248
> At5g18650
Length=267
Score = 72.8 bits (177), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 33/70 (47%), Positives = 47/70 (67%), Gaps = 1/70 (1%)
Query 48 CPVCSKSIADYSEFWKQLSDEIARTPMEDDMK-RKVRIACNDCLERSTTDFHFLGLKCMH 106
CP+CS+S+ D S+ W++L +EI T M D + +KV I CNDC + + FH +G KC H
Sbjct 192 CPICSRSVIDMSKTWQRLDEEIEATAMPSDYRDKKVWILCNDCNDTTEVHFHIIGQKCGH 251
Query 107 CNSFNTRDVA 116
C S+NTR +A
Sbjct 252 CRSYNTRAIA 261
> At5g25560
Length=272
Score = 68.2 bits (165), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 32/80 (40%), Positives = 49/80 (61%), Gaps = 1/80 (1%)
Query 35 RLLQKQKGFQAIRCPVCSKSIADYSEFWKQLSDEIARTPM-EDDMKRKVRIACNDCLERS 93
+ L++ + CP+CSKS+ D S+ W++ EIA TPM E R V+I CNDC +++
Sbjct 189 KCLEEMRDHYQYACPLCSKSVCDMSKVWEKFDMEIAATPMPEPYQNRMVQILCNDCGKKA 248
Query 94 TTDFHFLGLKCMHCNSFNTR 113
+H + KC +C S+NTR
Sbjct 249 EVQYHVVAQKCPNCKSYNTR 268
> 7297117
Length=496
Score = 67.4 bits (163), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 35/87 (40%), Positives = 48/87 (55%), Gaps = 6/87 (6%)
Query 32 DCLRLLQKQKGFQAI-----RCPVCSKSIADYSEFWKQLSDEIARTPMEDDMK-RKVRIA 85
DC LL K Q + CP C S+ D + W L D+ R P+ + ++V I
Sbjct 392 DCGHLLHKMCFDQLLASGHYTCPTCQTSLIDMTALWVYLDDQAERMPVPLKYENQRVHIF 451
Query 86 CNDCLERSTTDFHFLGLKCMHCNSFNT 112
CNDC + S T FHF+GLKC+HC ++NT
Sbjct 452 CNDCHKTSKTKFHFIGLKCVHCGAYNT 478
> At5g22920
Length=291
Score = 67.0 bits (162), Expect = 9e-12, Method: Compositional matrix adjust.
Identities = 33/69 (47%), Positives = 40/69 (57%), Gaps = 1/69 (1%)
Query 48 CPVCSKSIADYSEFWKQLSDEIARTPMEDDMKRK-VRIACNDCLERSTTDFHFLGLKCMH 106
CPVCSKSI D S WK+L +E+A PM + K V I CNDC + FH + KC
Sbjct 202 CPVCSKSICDMSNLWKKLDEEVAAYPMPKMYENKMVWILCNDCGSNTNVRFHLIAHKCSS 261
Query 107 CNSFNTRDV 115
C S+NTR
Sbjct 262 CGSYNTRQT 270
> SPAC2F3.16
Length=425
Score = 56.6 bits (135), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 29/67 (43%), Positives = 38/67 (56%), Gaps = 1/67 (1%)
Query 47 RCPVCSKSIADYSEFWKQLSDEIARTPMEDDMKRKVR-IACNDCLERSTTDFHFLGLKCM 105
RCP C K+I + + ++ L EI R PM + I CNDC R T +HFLG KC
Sbjct 308 RCPTCYKTIINVNSLFRILDMEIERQPMPYPYNTWISTIRCNDCNSRCDTKYHFLGHKCN 367
Query 106 HCNSFNT 112
C+S+NT
Sbjct 368 SCHSYNT 374
> At3g18290
Length=1254
Score = 55.1 bits (131), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 26/72 (36%), Positives = 42/72 (58%), Gaps = 1/72 (1%)
Query 48 CPVCSKSIADYSEFWKQLSDEIARTPMEDDMKRKVR-IACNDCLERSTTDFHFLGLKCMH 106
CP+C KS+ D + ++ L +A + ++ K + + I CNDC + TT FH+L KC
Sbjct 1173 CPICGKSLGDMAVYFGMLDALLAAEELPEEYKNRCQDILCNDCERKGTTRFHWLYHKCGS 1232
Query 107 CNSFNTRDVAED 118
C S+NTR + +
Sbjct 1233 CGSYNTRVIKSE 1244
> At1g74760
Length=255
Score = 55.1 bits (131), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 27/67 (40%), Positives = 38/67 (56%), Gaps = 1/67 (1%)
Query 48 CPVCSKSIADYSEFWKQLSDEIARTPMEDDMKRKVR-IACNDCLERSTTDFHFLGLKCMH 106
CPVCSKS+ D ++K L +A M D+ K + I CNDC + +H+L KC
Sbjct 187 CPVCSKSLGDMQVYFKMLDALLAEEKMPDEYSNKTQVILCNDCGRKGNAPYHWLYHKCTT 246
Query 107 CNSFNTR 113
C S+N+R
Sbjct 247 CGSYNSR 253
> At3g62970
Length=274
Score = 53.1 bits (126), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 30/85 (35%), Positives = 42/85 (49%), Gaps = 16/85 (18%)
Query 32 DCLRLLQKQKGFQAIRCPVCSKSIADYSEFWKQLSDEIARTPMEDDMKRKVRIACNDCLE 91
DC + + + RCP+C+KS+ D S W L E++ I CNDC +
Sbjct 196 DCFEQMINENQY---RCPICAKSMVDMSPSWHLLDFEVS-------------ILCNDCNK 239
Query 92 RSTTDFHFLGLKCMHCNSFNTRDVA 116
S FH LG KC C S+NTR ++
Sbjct 240 GSKAMFHILGHKCSDCGSYNTRRIS 264
> At1g18910
Length=195
Score = 52.0 bits (123), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 25/67 (37%), Positives = 38/67 (56%), Gaps = 1/67 (1%)
Query 48 CPVCSKSIADYSEFWKQLSDEIARTPMEDDMKRKVR-IACNDCLERSTTDFHFLGLKCMH 106
CP+CSKS+ D +++ L +A M D+ + + I CNDC + +H+L KC
Sbjct 127 CPICSKSLGDMQVYFRMLDALLAEQKMPDEYLNQTQVILCNDCGRKGNAPYHWLYHKCSS 186
Query 107 CNSFNTR 113
C S+NTR
Sbjct 187 CASYNTR 193
> Hs20555413
Length=1205
Score = 30.8 bits (68), Expect = 0.68, Method: Compositional matrix adjust.
Identities = 23/88 (26%), Positives = 38/88 (43%), Gaps = 9/88 (10%)
Query 26 LSLVAQDCLRLLQKQKGFQAIRCPVCSKSIADYSEFWKQLSDEIARTPME-DDMKRKVRI 84
+++ + L LLQ++ PV + DY EF + PM+ M+RK+
Sbjct 594 FNVLLRTTLDLLQEKDPAHIFAEPVNLSEVPDYLEFISK--------PMDFSTMRRKLES 645
Query 85 ACNDCLERSTTDFHFLGLKCMHCNSFNT 112
LE DF+ + CM N+ +T
Sbjct 646 HLYRTLEEFEEDFNLIVTNCMKYNAKDT 673
> Hs7657703
Length=1053
Score = 30.0 bits (66), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 23/88 (26%), Positives = 34/88 (38%), Gaps = 11/88 (12%)
Query 30 AQDCLRLLQKQKGFQAIRCPVCSKSIADYSEFWKQLSDEIARTPMEDDMKRKVRIACNDC 89
A+ R ++ G + CPVCS++ D K + T D + C
Sbjct 898 ARSLKRHVRTHTGERPYVCPVCSEAYIDARTLRKHM------TKFHRDY-----VPCKIM 946
Query 90 LERSTTDFHFLGLKCMHCNSFNTRDVAE 117
LE+ T FH G + H S T + E
Sbjct 947 LEKDTLQFHNQGTQVAHAVSILTAGMQE 974
> Hs17459695
Length=150
Score = 30.0 bits (66), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 15/52 (28%), Positives = 23/52 (44%), Gaps = 0/52 (0%)
Query 25 YLSLVAQDCLRLLQKQKGFQAIRCPVCSKSIADYSEFWKQLSDEIARTPMED 76
YLS + ++ G + +C C K A SE L D I R+P+ +
Sbjct 96 YLSFLQASKHAHIRSHAGKKPYKCDKCGKDFAKSSELKATLRDTIVRSPVSE 147
> CE20788
Length=548
Score = 28.9 bits (63), Expect = 2.7, Method: Composition-based stats.
Identities = 15/52 (28%), Positives = 24/52 (46%), Gaps = 3/52 (5%)
Query 47 RCPVCSKSIADYSEFWKQLSDEIARTPMEDDMKRKVRIACNDCLERSTTDFH 98
R P + I+ W SD +++ + DD + K+ C + S TDFH
Sbjct 234 RPPRFVQDISMAKRLW---SDVTSKSALSDDHRPKIEQICAAAMANSYTDFH 282
> SPCC736.11
Length=834
Score = 28.1 bits (61), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 20/76 (26%), Positives = 34/76 (44%), Gaps = 8/76 (10%)
Query 37 LQKQKGFQAIRCPVCSKSIADYSEFWKQLSDEIARTPMEDDMKRKVRIACNDCLERSTTD 96
++ +GF ++ + S S FW+ +D + + ME VR L+R +
Sbjct 187 IRPNQGFMSVNVDISS------SAFWR--NDSLLQILMEYTDCSNVRDLTRFDLKRLSRK 238
Query 97 FHFLGLKCMHCNSFNT 112
F FL + C H N+ T
Sbjct 239 FRFLKVTCQHRNNVGT 254
> Hs14747866
Length=1496
Score = 28.1 bits (61), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 11/29 (37%), Positives = 18/29 (62%), Gaps = 0/29 (0%)
Query 79 KRKVRIACNDCLERSTTDFHFLGLKCMHC 107
K +R+ C DCL+R+ T F+ L+ +H
Sbjct 379 KGTLRMNCLDCLDRTNTVQSFIALEVLHL 407
> Hs18584681
Length=427
Score = 27.3 bits (59), Expect = 6.6, Method: Compositional matrix adjust.
Identities = 11/45 (24%), Positives = 18/45 (40%), Gaps = 0/45 (0%)
Query 30 AQDCLRLLQKQKGFQAIRCPVCSKSIADYSEFWKQLSDEIARTPM 74
+ D ++ + G + +CP C K D S F +S P
Sbjct 247 SSDLIKHQRTHTGERPFKCPECGKGFRDSSHFVAHMSTHSGERPF 291
> 7304125
Length=429
Score = 27.3 bits (59), Expect = 7.1, Method: Composition-based stats.
Identities = 24/123 (19%), Positives = 51/123 (41%), Gaps = 17/123 (13%)
Query 1 PLHAGSCCNGLPSPEHQYRNYSLEYLSLVAQDCLRLLQK--------------QKGFQAI 46
P+ S C G P + +Y NY + + Q+ + L QK ++ Q +
Sbjct 14 PVAGDSGCRGNPPDQARYDNYRIYNVEFENQEQIELFQKLEEQSDSLTFIGHAREVGQKL 73
Query 47 RCPVCSKSIADYSEFWK--QLSDEIARTPMEDDMKRKVRIACNDCLERSTTDF-HFLGLK 103
V + +AD ++ K ++ + ++ + R + + ++ S D+ HF LK
Sbjct 74 SILVAAHRVADIADLLKTYKVKHRVLTYNFQEKIDRNLAEVQPESIDASQLDWQHFFHLK 133
Query 104 CMH 106
++
Sbjct 134 TIY 136
> ECU06g1270
Length=201
Score = 26.9 bits (58), Expect = 8.0, Method: Compositional matrix adjust.
Identities = 14/31 (45%), Positives = 19/31 (61%), Gaps = 1/31 (3%)
Query 38 QKQKGFQAIRCPVCSKSIADYSEFWKQLSDE 68
Q++K + R PV + A SE WK+LSDE
Sbjct 58 QRKKNEELSRLPVADQGRA-ISEMWKKLSDE 87
> CE08159
Length=413
Score = 26.9 bits (58), Expect = 8.4, Method: Compositional matrix adjust.
Identities = 13/31 (41%), Positives = 14/31 (45%), Gaps = 0/31 (0%)
Query 86 CNDCLERSTTDFHFLGLKCMHCNSFNTRDVA 116
C C T +HF CM C SF R VA
Sbjct 60 CQVCSTSERTSYHFGTTTCMACASFFRRTVA 90
> Hs14150005
Length=227
Score = 26.9 bits (58), Expect = 10.0, Method: Compositional matrix adjust.
Identities = 20/58 (34%), Positives = 25/58 (43%), Gaps = 4/58 (6%)
Query 35 RLLQKQKGFQAIRCPVCSKSIA-DYSEFWKQLSDEIARTPMEDDMKRKVRIACNDCLE 91
R GF +CP+CSKS+A D E + R DD+ K C CLE
Sbjct 135 RWFSSHSGF---KCPICSKSVASDEMEMHFIMCLSKPRLSYNDDVLTKDAGECVICLE 189
Lambda K H
0.323 0.135 0.438
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1167969826
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40