bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_1232_orf2
Length=484
Score E
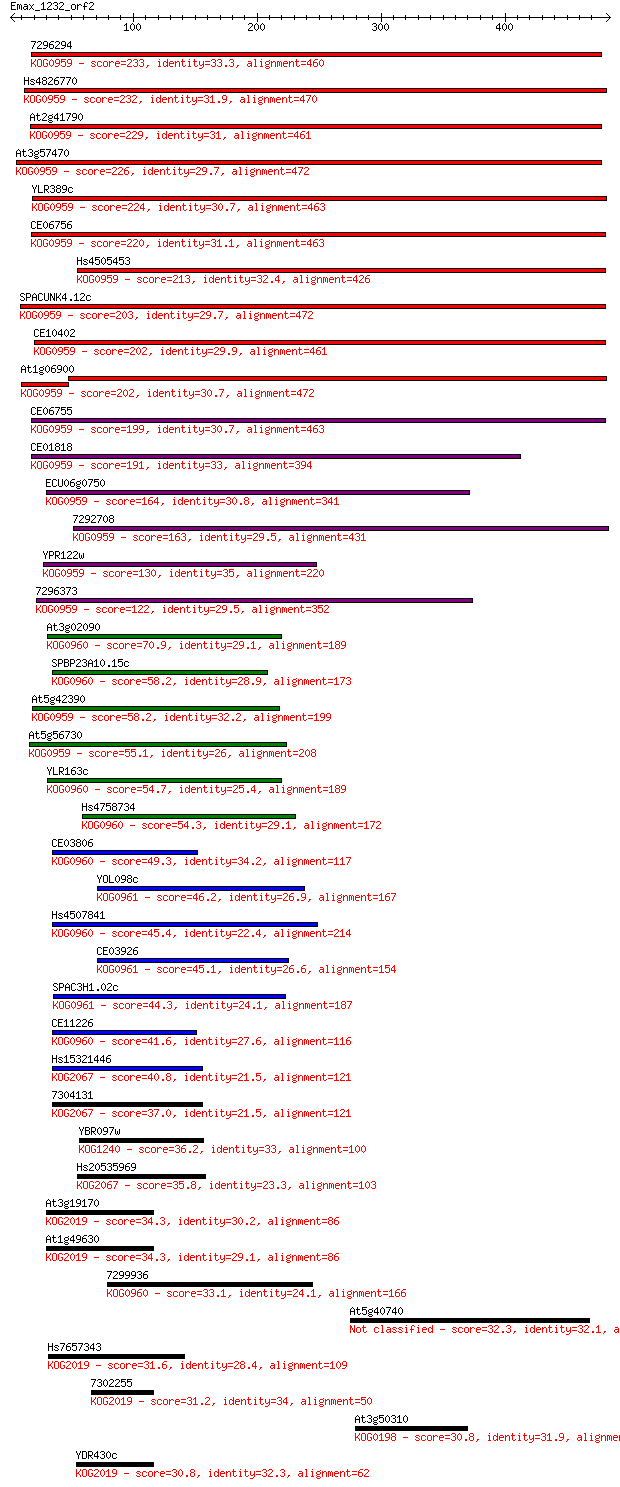
Sequences producing significant alignments: (Bits) Value
7296294 233 6e-61
Hs4826770 232 1e-60
At2g41790 229 8e-60
At3g57470 226 7e-59
YLR389c 224 2e-58
CE06756 220 5e-57
Hs4505453 213 1e-54
SPACUNK4.12c 203 6e-52
CE10402 202 2e-51
At1g06900 202 2e-51
CE06755 199 2e-50
CE01818 191 3e-48
ECU06g0750 164 5e-40
7292708 163 6e-40
YPR122w 130 7e-30
7296373 122 2e-27
At3g02090 70.9 6e-12
SPBP23A10.15c 58.2 4e-08
At5g42390 58.2 4e-08
At5g56730 55.1 3e-07
YLR163c 54.7 5e-07
Hs4758734 54.3 5e-07
CE03806 49.3 2e-05
YOL098c 46.2 2e-04
Hs4507841 45.4 3e-04
CE03926 45.1 4e-04
SPAC3H1.02c 44.3 5e-04
CE11226 41.6 0.004
Hs15321446 40.8 0.008
7304131 37.0 0.097
YBR097w 36.2 0.15
Hs20535969 35.8 0.25
At3g19170 34.3 0.60
At1g49630 34.3 0.70
7299936 33.1 1.2
At5g40740 32.3 2.2
Hs7657343 31.6 4.5
7302255 31.2 4.8
At3g50310 30.8 6.5
YDR430c 30.8 7.2
> 7296294
Length=990
Score = 233 bits (594), Expect = 6e-61, Method: Compositional matrix adjust.
Identities = 153/468 (32%), Positives = 234/468 (50%), Gaps = 25/468 (5%)
Query 18 NDILKPEADYRDFRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLE 77
N+I K D RD+R QL NG+ + + P ++ + A++ G + DP ++PGLAHF E
Sbjct 25 NNIEKSLQDTRDYRGLQLENGLKVLLISDPNTDVSAAALSVQVGHMSDPTNLPGLAHFCE 84
Query 78 HMLFLGTSKYPEPESYDGFLTQNGGANNAYTDEEKTVFFNKVTDSAFEEALDRFAEFFKS 137
HMLFLGT KYP Y +L+Q+GG++NA T T + V + ALDRFA+FF +
Sbjct 85 HMLFLGTEKYPHENGYTTYLSQSGGSSNAATYPLMTKYHFHVAPDKLDGALDRFAQFFIA 144
Query 138 PLFSRQYEEKEVNAIDAEHQKNIPNDDERAWYTIRSLAK--GPMSRFATGNSETLSTNPK 195
PLF+ E+E+NA+++EH+KN+P+D R R LAK S+F +GN TLS PK
Sbjct 145 PLFTPSATEREINAVNSEHEKNLPSDLWRIKQVNRHLAKPDHAYSKFGSGNKTTLSEIPK 204
Query 196 AKGIDLVDRLRDFHSKYYCGSNMVAVTISPRSLDEQESLIREKLEGISAGHADWLGMVQC 255
+K ID+ D L FH ++Y + M I SLDE E ++ EK I + V+
Sbjct 205 SKNIDVRDELLKFHKQWYSANIMCLAVIGKESLDELEGMVLEKFSEIENKN------VKV 258
Query 256 PG-PMFDTVKPFDHTNSGKFIHMQSFSSEPSLWVAFGLPATLTSYKKQPTSVLTYLLEYT 314
PG P P+ G+ + + SL ++F YK P + LT+L+ +
Sbjct 259 PGWPR----HPYAEERYGQKVKIVPIKDIRSLTISFTTDDLTQFYKSGPDNYLTHLIGHE 314
Query 315 GEGSLAKRLRLLGLADGISPAVDRNSIS--TLLGIKVDLTQKGAAHRGLVLQEIFSYINF 372
G+GS+ LR LG + + A +N+ + I VDLTQ+G H +++ +F Y+
Sbjct 315 GKGSILSELRRLGWCNDLM-AGHQNTQNGFGFFDIVVDLTQEGLEHVDDIVKIVFQYLEM 373
Query 373 LRDHGVGHELVSTMAQQSHIDFH---TTQPSSSIMDEAARLAHNLLTYEPYHVVAGDSLL 429
LR G + + + + F QP + + + + + V+ L
Sbjct 374 LRKEGPKKWIFDECVKLNEMRFRFKEKEQPENLVTHAVSSMQ----IFPLEEVLIAPYLS 429
Query 430 IDADPRLTNQLLQEMSPSKAIIAFSDPDFTSKVDSFETDPYYGVQFRV 477
+ P L LL E+ PSK+ I F D E PYY ++ +
Sbjct 430 NEWRPDLIKGLLDELVPSKSRIVIVSQSFEPDCDLAE--PYYKTKYGI 475
> Hs4826770
Length=1019
Score = 232 bits (591), Expect = 1e-60, Method: Compositional matrix adjust.
Identities = 150/474 (31%), Positives = 237/474 (50%), Gaps = 17/474 (3%)
Query 12 AIGALGNDILKPEADYRDFRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPG 71
AI +GN I K D R++R +L+NG+ + + P ++++ A+ + GSL DP ++ G
Sbjct 46 AIKRIGNHITKSPEDKREYRGLELANGIKVLLMSDPTTDKSSAALDVHIGSLSDPPNIAG 105
Query 72 LAHFLEHMLFLGTSKYPEPESYDGFLTQNGGANNAYTDEEKTVFFNKVTDSAFEEALDRF 131
L+HF EHMLFLGT KYP+ Y FL+++ G++NA+T E T ++ V+ E ALDRF
Sbjct 106 LSHFCEHMLFLGTKKYPKENEYSQFLSEHAGSSNAFTSGEHTNYYFDVSHEHLEGALDRF 165
Query 132 AEFFKSPLFSRQYEEKEVNAIDAEHQKNIPNDDERAWYTIRSLA--KGPMSRFATGNSET 189
A+FF PLF +++EVNA+D+EH+KN+ ND R + ++ K P S+F TGN T
Sbjct 166 AQFFLCPLFDESCKDREVNAVDSEHEKNVMNDAWRLFQLEKATGNPKHPFSKFGTGNKYT 225
Query 190 LSTNPKAKGIDLVDRLRDFHSKYYCGSNMVAVTISPR-SLDEQESLIREKLEGISAGHAD 248
L T P +GID+ L FHS YY SN++AV + R SLD+ +L+ + + +
Sbjct 226 LETRPNQEGIDVRQELLKFHSAYY-SSNLMAVCVLGRESLDDLTNLVVKLFSEVENKNV- 283
Query 249 WLGMVQCPGPMFDTVKPFDHTNSGKFIHMQSFSSEPSLWVAFGLPATLTSYKKQPTSVLT 308
P P F PF + + + +L+V F +P YK P L
Sbjct 284 -------PLPEFPE-HPFQEEHLKQLYKIVPIKDIRNLYVTFPIPDLQKYYKSNPGHYLG 335
Query 309 YLLEYTGEGSLAKRLRLLGLADGISPAVDRNSISTLLG-IKVDLTQKGAAHRGLVLQEIF 367
+L+ + G GSL L+ G + + + + I VDLT++G H ++ +F
Sbjct 336 HLIGHEGPGSLLSELKSKGWVNTLVGGQKEGARGFMFFIINVDLTEEGLLHVEDIILHMF 395
Query 368 SYINFLRDHGVGHELVSTMAQQSHIDFHTTQPSSSIMDEAARLAHNLLTYEPYHVVAGDS 427
YI LR G + + + F +++A L Y V+ +
Sbjct 396 QYIQKLRAEGPQEWVFQECKDLNAVAFRFKDKERP-RGYTSKIAGILHYYPLEEVLTAEY 454
Query 428 LLIDADPRLTNQLLQEMSPSKAIIAFSDPDFTSKVDSFETDPYYGVQFRVLDLP 481
LL + P L +L ++ P +A F K D T+ +YG Q++ +P
Sbjct 455 LLEEFRPDLIEMVLDKLRPENVRVAIVSKSFEGKTD--RTEEWYGTQYKQEAIP 506
> At2g41790
Length=970
Score = 229 bits (585), Expect = 8e-60, Method: Compositional matrix adjust.
Identities = 143/467 (30%), Positives = 236/467 (50%), Gaps = 21/467 (4%)
Query 17 GNDILKPEADYRDFRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFL 76
G +ILKP D R++R L N + + + P +++ +++ + GS DP+ + GLAHFL
Sbjct 12 GVEILKPRTDNREYRMIVLKNLLQVLLISDPDTDKCAASMSVSVGSFSDPQGLEGLAHFL 71
Query 77 EHMLFLGTSKYPEPESYDGFLTQNGGANNAYTDEEKTVFFNKVTDSAFEEALDRFAEFFK 136
EHMLF + KYPE +SY ++T++GG+ NAYT E+T + V F+EALDRFA+FF
Sbjct 72 EHMLFYASEKYPEEDSYSKYITEHGGSTNAYTASEETNYHFDVNADCFDEALDRFAQFFI 131
Query 137 SPLFSRQYEEKEVNAIDAEHQKNIPNDDERAWYTIRSLAKG--PMSRFATGNSETLSTNP 194
PL S +E+ A+D+E+QKN+ +D R + L+K P +F+TGN +TL P
Sbjct 132 KPLMSADATMREIKAVDSENQKNLLSDGWRIRQLQKHLSKEDHPYHKFSTGNMDTLHVRP 191
Query 195 KAKGIDLVDRLRDFHSKYYCGSNMVAVTISPRSLDEQESLIREKLEGISAGHADWLGMVQ 254
+AKG+D L F+ ++Y + M V SLD+ + L+ + I + + +
Sbjct 192 QAKGVDTRSELIKFYEEHYSANIMHLVVYGKESLDKIQDLVERMFQEIQNTNK---VVPR 248
Query 255 CPGPMFDTVKPFDHTNSGKFIHMQSFSSEPSLWVAFGLPATLTSYKKQPTSVLTYLLEYT 314
PG +P + + L V++ + ++ Y + P+ L +L+ +
Sbjct 249 FPG------QPCTADHLQILVKAIPIKQGHKLGVSWPVTPSIHHYDEAPSQYLGHLIGHE 302
Query 315 GEGSLAKRLRLLGLADGISPAVDRNSIS-TLLGIKVDLTQKGAAHRGLVLQEIFSYINFL 373
GEGSL L+ LG A G+S ++ + + +DLT G H +L +F+YI L
Sbjct 303 GEGSLFHALKTLGWATGLSAGEGEWTLDYSFFKVSIDLTDAGHEHMQEILGLLFNYIQLL 362
Query 374 RDHGVGHELVSTMAQQSHIDFH---TTQPSSSIMDEAARLAHNLLTYEPYHVVAGDSLLI 430
+ GV + ++ FH P S I+D +A N+ Y + G SL
Sbjct 363 QQTGVCQWIFDELSAICETKFHYQDKIPPMSYIVD----IASNMQIYPTKDWLVGSSLPT 418
Query 431 DADPRLTNQLLQEMSPSKAIIAFSDPDFTSKVDSFETDPYYGVQFRV 477
+P + +++ E+SPS I + F + D E P+Y + +
Sbjct 419 KFNPAIVQKVVDELSPSNFRIFWESQKFEGQTDKAE--PWYNTAYSL 463
> At3g57470
Length=989
Score = 226 bits (577), Expect = 7e-59, Method: Compositional matrix adjust.
Identities = 140/489 (28%), Positives = 240/489 (49%), Gaps = 27/489 (5%)
Query 6 LCVGLSAIGALGN--DILKPEADYRDFRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSL 63
+ VG+ A G +ILKP D R++R L N + + + P +++ ++ + GS
Sbjct 1 MAVGMENATASGECGEILKPRTDKREYRRIVLKNSLEVLLISDPETDKCAASMNVSVGSF 60
Query 64 YDPEDVPGLAHFLEHMLFLGTSKYPEPESYDGFLTQNGGANNAYTDEEKTVFFNKVTDSA 123
DPE + GLAHFLEHMLF + KYPE +SY ++T++GG+ NAYT E T + + +
Sbjct 61 TDPEGLEGLAHFLEHMLFYASEKYPEEDSYSKYITEHGGSTNAYTSSEDTNYHFDINTDS 120
Query 124 FEEALDRFAEFFKSPLFSRQYEEKEVNAIDAEHQKNIPNDDERAWYTIRSLAKG--PMSR 181
F EALDRFA+FF PL S +E+ A+D+EHQ N+ +D R + L++ P +
Sbjct 121 FYEALDRFAQFFIQPLMSTDATMREIKAVDSEHQNNLLSDSWRMAQLQKHLSREDHPYHK 180
Query 182 FATGNSETLSTNPKAKGIDLVDRLRDFHSKYYCGSNMVAVTISPRSLDEQESLIREKLEG 241
F+TGN +TL P+ G+D L F+ ++Y + M V +LD+ + L+ +G
Sbjct 181 FSTGNMDTLHVRPEENGVDTRSELIKFYDEHYSANIMHLVVYGKENLDKTQGLVEALFQG 240
Query 242 ISAGHADWLGMVQCPGPMFDTVKPFDHTNSGKFIHMQSFSSEPSLWVAFGLPATLTSYKK 301
I + G+ + PG +P + + L V++ + +++ Y++
Sbjct 241 IRNTNQ---GIPRFPG------QPCTLDHLQVLVKAVPIMQGHELSVSWPVTPSISHYEE 291
Query 302 QPTSVLTYLLEYTGEGSLAKRLRLLGLADGI-SPAVDRNSISTLLGIKVDLTQKGAAHRG 360
P L L+ + GEGSL L++LG A G+ + D + + + +DLT G H
Sbjct 292 APCRYLGDLIGHEGEGSLFHALKILGWATGLYAGEADWSMEYSFFNVSIDLTDAGHEHMQ 351
Query 361 LVLQEIFSYINFLRDHGVGHELVSTMAQQSHIDFHTTQPSSSIMDEAARLAHNLLTYEPY 420
+L +F YI L+ GV + ++ +FH Q + A ++ N+ Y
Sbjct 352 DILGLLFEYIKVLQQSGVSQWIFDELSAICEAEFH-YQAKIDPISYAVDISSNMKIYPTK 410
Query 421 HVVAGDSLLIDADPRLTNQLLQEMSPSKA------------IIAFSDPDFTSKVDSFETD 468
H + G SL +P + ++L E+SP+ ++ +PD + + +
Sbjct 411 HWLVGSSLPSKFNPAIVQKVLDELSPNNPSVPNVLCSYNLHVVQALNPDCLRPRQTDKVE 470
Query 469 PYYGVQFRV 477
P+Y + +
Sbjct 471 PWYNTAYSL 479
> YLR389c
Length=988
Score = 224 bits (572), Expect = 2e-58, Method: Compositional matrix adjust.
Identities = 142/473 (30%), Positives = 245/473 (51%), Gaps = 30/473 (6%)
Query 19 DILKPEADYRDFRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEH 78
D LKP+ D R +R +L N + A+ + P++++A ++ N G+ DP+++PGLAHF EH
Sbjct 63 DFLKPDLDERSYRFIELPNKLKALLIQDPKADKAAASLDVNIGAFEDPKNLPGLAHFCEH 122
Query 79 MLFLGTSKYPEPESYDGFLTQNGGANNAYTDEEKTVFFNKVTDSAFEEALDRFAEFFKSP 138
+LF+G+ K+P+ Y +L+++GG++NAYT + T +F +V ALDRF+ FF P
Sbjct 123 LLFMGSEKFPDENEYSSYLSKHGGSSNAYTASQNTNYFFEVNHQHLFGALDRFSGFFSCP 182
Query 139 LFSRQYEEKEVNAIDAEHQKNIPNDDERAWYTIRSL--AKGPMSRFATGNSETLSTNPKA 196
LF++ +KE+NA+++E++KN+ ND R + +SL K P +F+TGN ETL T PK
Sbjct 183 LFNKDSTDKEINAVNSENKKNLQNDIWRIYQLDKSLTNTKHPYHKFSTGNIETLGTLPKE 242
Query 197 KGIDLVDRLRDFHSKYYCGSNMVAVTISPRSLDEQESLIREKLEGISAGHADWLGMVQCP 256
G+++ D L FH +Y +N++ + I L RE L+ +S D V
Sbjct 243 NGLNVRDELLKFHKNFY-SANLMKLCI----------LGREDLDTLSDWTYDLFKDVANN 291
Query 257 G---PMF-DTVKPFDHTNSGKFIHMQSFSSEPSLWVAFGLPATLTSYKKQPTSVLTYLLE 312
G P++ + + +H K I ++ L ++F +P ++ +P +L++L+
Sbjct 292 GREVPLYAEPIMQPEHLQ--KIIQVRPVKDLKKLEISFTVPDMEEHWESKPPRILSHLIG 349
Query 313 YTGEGSLAKRLRLLGLADGISPAVDRNSI-STLLGIKVDLTQKGAAHRGLVLQEIFSYIN 371
+ G GSL L+ LG A+ +S S + + +DLT G H V+ IF YI
Sbjct 350 HEGSGSLLAHLKKLGWANELSAGGHTVSKGNAFFAVDIDLTDNGLTHYRDVIVLIFQYIE 409
Query 372 FLRDHGVGHELVSTMAQQSHIDFHTTQ---PSSSIMDEAARLAHNLLTYEPYHVVAGDSL 428
L++ + + + S+ F Q PSS++ A L + Y P + L
Sbjct 410 MLKNSLPQKWIFNELQDISNATFKFKQAGSPSSTVSSLAKCLEKD---YIPVSRILAMGL 466
Query 429 LIDADPRLTNQLLQEMSPSKAIIAFSDPDFTSKVDSFETDPYYGVQFRVLDLP 481
L +P L Q + P + + + +++ + +YG ++V+D P
Sbjct 467 LTKYEPDLLTQYTDALVPENSRVTL----ISRSLETDSAEKWYGTAYKVVDYP 515
> CE06756
Length=845
Score = 220 bits (561), Expect = 5e-57, Method: Compositional matrix adjust.
Identities = 144/473 (30%), Positives = 236/473 (49%), Gaps = 27/473 (5%)
Query 18 NDILKPEADYRDFRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLE 77
N I+K D R+ R +L+NG+ + V P ++++ ++A G L DP ++PGLAHF E
Sbjct 14 NSIVKGPQDERECRGLELTNGLRVLLVSDPTTDKSAVSLAVKAGHLMDPWELPGLAHFCE 73
Query 78 HMLFLGTSKYPEPESYDGFLTQNGGANNAYTDEEKTVFFNKVTDSAFEEALDRFAEFFKS 137
HMLFLGTSKYP + FL+ N G+ NA T+ + T + V ALDRF +FF
Sbjct 74 HMLFLGTSKYPLENEFTKFLSDNAGSYNACTEPDHTYYHFDVKPDQLYGALDRFVQFFLC 133
Query 138 PLFSRQYEEKEVNAIDAEHQKNIPNDDERAWYTIRSLAK-GPMSR-FATGNSETLSTNPK 195
P F++ E+EV A+D+EH N+ +D R RSL++ G +R F TGN +TL + +
Sbjct 134 PQFTKSATEREVCAVDSEHLSNLNSDYWRILQVDRSLSRPGHDNRKFCTGNKKTLLEDAR 193
Query 196 AKGIDLVDRLRDFHSKYYCGSNMVAVTISPRSLDEQESLIR----EKLEGISAGHADWLG 251
KGI+ D L +F+ K+Y + M I SLD ES +R + ++ A W
Sbjct 194 KKGIEPRDALLEFYKKWYSSNIMTCCIIGKESLDVLESYLRTLEFDAIQNKKAESKVWAE 253
Query 252 MVQCPGPMFDTVKPFDHTNSGKFIHMQSFSSEPSLWVAFGLPATLTSYKKQPTSVLTYLL 311
P + K I + + + + F P Y QP + +L+
Sbjct 254 FQYGPDQL------------AKKIDVVPIKDKKLVSIIFPFPDLNNEYLSQPGHYIAHLI 301
Query 312 EYTGEGSLAKRLRLLGLADGISPAVDRNSISTLLG---IKVDLTQKGAAHRGLVLQEIFS 368
+ G GS++ L+ LG A + P + +I+ G + +DL+ +G H ++Q +F+
Sbjct 302 GHKGPGSISSELKRLGWASSLKP--ESKTIAAGFGYFNVTMDLSTEGLEHVDEIIQLMFN 359
Query 369 YINFLRDHGVGHELVSTMAQQSHIDFHTTQPSSSIMDEAARLAHNLLTYEPY-HVVAGDS 427
YI L+ G + +A+ S I+F + + A ++A N L Y P+ H+++
Sbjct 360 YIGMLQSAGPQQWIHEELAELSAIEFR-FKDREPLTKNAIKVARN-LQYIPFEHILSSRY 417
Query 428 LLIDADPRLTNQLLQEMSPSKAIIAFSDPDFTSKVDSFETDPYYGVQFRVLDL 480
LL +P +LL ++PS ++ F + + +P YG + +V D+
Sbjct 418 LLTKYNPERIKELLSTLTPSNMLVRVVSKKFKEQ-EGNTNEPVYGTEMKVTDI 469
> Hs4505453
Length=1219
Score = 213 bits (541), Expect = 1e-54, Method: Compositional matrix adjust.
Identities = 138/434 (31%), Positives = 214/434 (49%), Gaps = 19/434 (4%)
Query 55 AVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYPEPESYDGFLTQNGGANNAYTDEEKTV 114
A+ GS DP+D+PGLAHFLEHM+F+G+ KYP+ +D FL ++GG++NA TD E+TV
Sbjct 282 ALCVGVGSFADPDDLPGLAHFLEHMVFMGSLKYPDENGFDAFLKKHGGSDNASTDCERTV 341
Query 115 FFNKVTDSAFEEALDRFAEFFKSPLFSRQYEEKEVNAIDAEHQKNIPNDDERAWYTIRSL 174
F V F+EALDR+A+FF PL R ++EV A+D+E+Q P+D R SL
Sbjct 342 FQFDVQRKYFKEALDRWAQFFIHPLMIRDAIDREVEAVDSEYQLARPSDANRKEMLFGSL 401
Query 175 AKG--PMSRFATGNSETLSTNPKAKGIDLVDRLRDFHSKYYCGSNMVAVTISPRSLDEQE 232
A+ PM +F GN+ETL P+ ID RLR+F +YY M V S +LD E
Sbjct 402 ARPGHPMGKFFWGNAETLKHEPRKNNIDTHARLREFWMRYYSSHYMTLVVQSKETLDTLE 461
Query 233 SLIREKLEGISAGHADWLGMVQCPGPMFDTVK-PFDHTNSGKFIHMQSFSSEPSLWVAFG 291
+ E I G+ P P F + PFD K + +L + +
Sbjct 462 KWVTEIFSQIPNN-----GL---PRPNFGHLTDPFDTPAFNKLYRVVPIRKIHALTITWA 513
Query 292 LPATLTSYKKQPTSVLTYLLEYTGEGS----LAKRLRLLGLADGI-SPAVDRNSISTLLG 346
LP Y+ +P +++L+ + G+GS L K+ L L G ++NS ++
Sbjct 514 LPPQQQHYRVKPLHYISWLVGHEGKGSILSFLRKKCWALALFGGNGETGFEQNSTYSVFS 573
Query 347 IKVDLTQKGAAHRGLVLQEIFSYINFLRDHGVGHELVSTMAQQSHIDFHTTQPSSSIMDE 406
I + LT +G H V +F Y+ L+ G + + + +FH Q + ++
Sbjct 574 ISITLTDEGYEHFYEVAYTVFLYLKMLQKLGPEKRIFEEIRKIEDNEFH-YQEQTDPVEY 632
Query 407 AARLAHNLLTYEPYHVVAGDSLLIDADPRLTNQLLQEMSPSKAIIAFSDPDFTSKVDSFE 466
+ N+ Y ++ GD LL + P + + L ++ P KA + K D E
Sbjct 633 VENMCENMQLYPLQDILTGDQLLFEYKPEVIGEALNQLVPQKANLVLLSGANEGKCDLKE 692
Query 467 TDPYYGVQFRVLDL 480
++G Q+ + D+
Sbjct 693 K--WFGTQYSIEDI 704
> SPACUNK4.12c
Length=969
Score = 203 bits (517), Expect = 6e-52, Method: Compositional matrix adjust.
Identities = 140/480 (29%), Positives = 226/480 (47%), Gaps = 24/480 (5%)
Query 9 GLSAIGALGNDILKPEADYRDFRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPED 68
G + + N + P D R++R +L N + + V P ++ A A+ + GS +P +
Sbjct 3 GKPQVEVIVNGQVVPNLDDREYRLIKLENDLEVLLVRDPETDNASAAIDVHIGSQSNPRE 62
Query 69 VPGLAHFLEHMLFLGTSKYPEPESYDGFLTQNGGANNAYTDEEKTVFFNKVTDSAFEEAL 128
+ GLAHF EH+LF+GT KYP+ Y +L + G +NAYT T ++ +V+ A AL
Sbjct 63 LLGLAHFCEHLLFMGTKKYPDENEYRKYLESHNGISNAYTASNNTNYYFEVSHDALYGAL 122
Query 129 DRFAEFFKSPLFSRQYEEKEVNAIDAEHQKNIPNDDERAW--YTIRSLAKGPMSRFATGN 186
DRFA+FF PLF + +++E+ A+D+EH KN+ +D R W Y++ S K S+F TGN
Sbjct 123 DRFAQFFIDPLFLEECKDREIRAVDSEHCKNLQSDSWRFWRLYSVLSNPKSVFSKFNTGN 182
Query 187 SETLSTNPKAKGIDLVDRLRDFHSKYYCGSNMVAVTISPRSLDEQESLIREKLEGISAGH 246
ETL PK G+D+ L F+ KYY + M V I LD + E I
Sbjct 183 IETLGDVPKELGLDVRQELLKFYDKYYSANIMKLVIIGREPLDVLQDWAAELFSPIKNK- 241
Query 247 ADWLGMVQCPGPMFDTVKPFDHTNSGKFIHMQSFSSEPSLWVAFGLPATLTSYKKQPTSV 306
P P F P+ K +++ + L + F +P YK +P
Sbjct 242 -------AVPIPKFPD-PPYTDNEVRKICYVKPVKNLRRLDIVFPIPGQYHKYKCRPAEY 293
Query 307 LTYLLEYTGEGSLAKRLRLLGLADG-ISPAVDRNSISTLLGIKVDLTQKGAAHRGLVLQE 365
+ +LL + GEGS L+ LGLA I+ V + ++ + LT++G V++
Sbjct 294 VCHLLGHEGEGSYLAYLKSLGLATSLIAFNVSITEDADIIVVSTFLTEEGLTDYQRVIKI 353
Query 366 IFSYINFLRDHGVGHELVSTMAQQSHIDFHTTQPSSSIMDEAARLAHNLLT-----YEPY 420
+F YI L L S F T Q + A + AH + + Y
Sbjct 354 LFEYIRLLDQTNAHKFLFEETRIMSEAQFKTRQKTP-----AYQYAHVVASKLQREYPRD 408
Query 421 HVVAGDSLLIDADPRLTNQLLQEMSPSKAIIAFSDPDFTSKVDSFETDPYYGVQFRVLDL 480
V+ S+L + DP+ ++++ + P+ + +D+ E +YG+ + + DL
Sbjct 409 KVLYYSSVLTEFDPKGIQEVVESLRPNNFFAILAAHSIEKGLDNKEK--FYGIDYGLEDL 466
> CE10402
Length=1067
Score = 202 bits (513), Expect = 2e-51, Method: Compositional matrix adjust.
Identities = 138/474 (29%), Positives = 223/474 (47%), Gaps = 33/474 (6%)
Query 20 ILKPEADYRDFRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHM 79
I+K D R++R +L+NG+ + V P ++++ A+ G L DP ++PGLAHF EHM
Sbjct 75 IVKGAQDAREYRGLELTNGIRVLLVSDPTTDKSAAALDVKVGHLMDPWELPGLAHFCEHM 134
Query 80 LFLGTSKYPEPESYDGFLTQNGGANNAYTDEEKTVFFNKVTDSAFEEALDRFAEFFKSPL 139
LFLGT+KYP Y FL + G++NAYT + T + V ALDRF +FF SP
Sbjct 135 LFLGTAKYPSENEYSKFLAAHAGSSNAYTSSDHTNYHFDVKPDQLPGALDRFVQFFLSPQ 194
Query 140 FSRQYEEKEVNAIDAEHQKNIPNDDERAWYTIRSLAK--GPMSRFATGNSETLSTNPKAK 197
F+ E+EV A+D+EH N+ ND R RS +K +F TGN +TL + + K
Sbjct 195 FTESATEREVCAVDSEHSNNLNNDLWRFLQVDRSRSKPGHDYGKFGTGNKQTLLEDARKK 254
Query 198 GIDLVDRLRDFHSKYYCGSNMVAVTISPRSLDEQESLIR----EKLEGISAGHADWLGMV 253
GI+ D L FH K+Y M + L+ ES + + +E W
Sbjct 255 GIEPRDALLQFHKKWYSSDIMTCCIVGKEPLNVLESYLGTLEFDAIENKKVERKVWEEF- 313
Query 254 QCPGPMFDTVKPFDHTNSGKFIHMQSFSSEPSLWVAFGLPATLTSYKKQPTSVLTYLLEY 313
P+ K I + + ++F P + QP +++L+ +
Sbjct 314 -----------PYGPDQLAKRIDVVPIKDTRLVSISFPFPDLNGEFLSQPGHYISHLIGH 362
Query 314 TGEGSLAKRLRLLGLADGISPAVDRNSISTLLG---IKVDLTQKGAAHRGLVLQEIFSYI 370
G GSL L+ LG + D ++ + G + +DL+ +G H ++Q +F+YI
Sbjct 363 EGPGSLLSELKRLGWVSSLQS--DSHTQAAGFGVYNVTMDLSTEGLEHVDEIIQLMFNYI 420
Query 371 NFLRDHGVGHELVSTMAQQSHIDFH---TTQPSSSIMDEAARLAHNLLTYEPY-HVVAGD 426
L+ G + +A+ S + F QP + ++ AA L Y P+ H+++
Sbjct 421 GMLQSAGPKQWVHDELAELSAVKFRFKDKEQPMTMAINVAAS-----LQYIPFEHILSSR 475
Query 427 SLLIDADPRLTNQLLQEMSPSKAIIAFSDPDFTSKVDSFETDPYYGVQFRVLDL 480
LL +P +LL +SP+ + F + + +P YG + +V D+
Sbjct 476 YLLTKYEPERIKELLSMLSPANMQVRVVSQKFKGQ-EGNTNEPVYGTEMKVTDI 528
> At1g06900
Length=1023
Score = 202 bits (513), Expect = 2e-51, Method: Compositional matrix adjust.
Identities = 132/443 (29%), Positives = 221/443 (49%), Gaps = 24/443 (5%)
Query 48 RSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYPEPESYDGFLTQNGGANNAY 107
++ +A A+ + GS DP + GLAHFLEHMLF+G++++P+ YD +L+++GG++NAY
Sbjct 103 QTKKAAAAMCVSMGSFLDPPEAQGLAHFLEHMLFMGSTEFPDENEYDSYLSKHGGSSNAY 162
Query 108 TDEEKTVFFNKVTDSAFEEALDRFAEFFKSPLFSRQYEEKEVNAIDAEHQKNIPNDDER- 166
T+ E T + +V + AL RF++FF +PL + E+EV A+D+E + + ND R
Sbjct 163 TEMEHTCYHFEVKREFLQGALKRFSQFFVAPLMKTEAMEREVLAVDSEFNQALQNDACRL 222
Query 167 AWYTIRSLAKG-PMSRFATGNSETLSTNPKAKGIDLVDRLRDFHSKYYCGSNMVAVTISP 225
+ AKG P +RFA G + + G+DL + + + +YY G M V I
Sbjct 223 QQLQCYTSAKGHPFNRFAWGKDTAVLSGAMENGVDLRECIVKLYKEYYHGGLMKLVVIGG 282
Query 226 RSLDEQESLIREKLEGISAGHADWLGMVQCPGPMFDTVKPFDHTNSGKFIHMQSFSSEPS 285
SLD ES + E + G + ++ GP++ GK +++
Sbjct 283 ESLDMLESWVVELFGDVKNG-SKIRPTLEAEGPIW---------KGGKLYRLEAVKDVHI 332
Query 286 LWVAFGLPATLTSYKKQPTSVLTYLLEYTGEGSLAKRLRLLGLADGISPAV-----DRNS 340
L + + LP ++Y K+P L +LL + G GSL L+ G A +S V +R+S
Sbjct 333 LDLTWTLPPLRSAYVKKPEDYLAHLLGHEGRGSLHSFLKAKGWATSLSAGVGDDGINRSS 392
Query 341 ISTLLGIKVDLTQKGAAHRGLVLQEIFSYINFLRDHGVGHELVSTMAQQSHIDFH--TTQ 398
++ + G+ + LT G ++ I+ Y+ LRD + + ++DF Q
Sbjct 393 LAYVFGMSIHLTDSGLEKIYDIIGYIYQYLKLLRDVSPQEWIFKELQDIGNMDFRFAEEQ 452
Query 399 PSSSIMDEAARLAHNLLTYEPYHVVAGDSLLIDADPRLTNQLLQEMSPSKAIIAFSDPDF 458
P+ D AA L+ N+L Y HV+ GD + DP+L L+ +P I
Sbjct 453 PAD---DYAAELSENMLAYPVEHVIYGDYVYQTWDPKLIEDLMGFFTPQNMRIDVVSKSI 509
Query 459 TSKVDSFETDPYYGVQFRVLDLP 481
S + F+ +P++G + D+P
Sbjct 510 KS--EEFQQEPWFGSSYIEEDVP 530
Score = 32.7 bits (73), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 13/38 (34%), Positives = 23/38 (60%), Gaps = 0/38 (0%)
Query 10 LSAIGALGNDILKPEADYRDFRHYQLSNGMHAIAVHHP 47
+ ++ AL N ++K D R +R +L NG+ A+ +H P
Sbjct 4 MKSVSALDNVVVKSPNDRRLYRVIELENGLCALLIHDP 41
> CE06755
Length=980
Score = 199 bits (505), Expect = 2e-50, Method: Compositional matrix adjust.
Identities = 142/472 (30%), Positives = 225/472 (47%), Gaps = 52/472 (11%)
Query 18 NDILKPEADYRDFRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLE 77
+ I+K D R++R +L+NG+ + V P ++++ A+ G L DP ++PGLAHF E
Sbjct 14 DSIVKGAQDDREYRGLELTNGIRVLLVSDPTTDKSAAALDVKVGHLMDPWELPGLAHFCE 73
Query 78 HMLFLGTSKYPEPESYDGFLTQNGGANNAYTDEEKTVFFNKVTDSAFEEALDRFAEFFKS 137
HMLFLGT+KYP Y FLT N G NA T + T + V ALDRF +FF S
Sbjct 74 HMLFLGTAKYPTENEYSKFLTDNAGHRNAVTASDHTNYHFDVKPDQLRGALDRFVQFFLS 133
Query 138 PLFSRQYEEKEVNAIDAEHQKNIPNDDERAWYTIRSLAK--GPMSRFATGNSETLSTNPK 195
P F+ E+EV A+D+EH N+ ND R RSL+K ++F TGN +TL +
Sbjct 134 PQFTESATEREVCAVDSEHSNNLNNDLWRLSQVDRSLSKPGHDYAKFGTGNKKTLLEEAR 193
Query 196 AKGIDLVDRLRDFHSKYYCGSNMVAVTISPRSLDEQESLIREKLEGISAGHADWLGMVQC 255
KG++ D L F+ K+Y + M I SLD +S ++ LE
Sbjct 194 KKGVEPRDALLQFYKKWYSSNIMTCCIIGKESLDVLQSHLK-TLE--------------- 237
Query 256 PGPMFDTVKPFDHTNSGKFIHMQSFSSEPSLWVAFGLPATLTSYKKQPTSVLTYLLEYTG 315
FDT++ K + + ++ P +G P L K+ +L+ + G
Sbjct 238 ----FDTIE-------NKKVERKVWNENP-----YG-PEQLG--KRIDRKFFAHLIRHKG 278
Query 316 EGSLAKRLRLLGLADGISPAVDRNSIST---LLGIKVDLTQKGAAHRGLVLQEIFSYINF 372
GSL L+ LG + + D N+I+ +L + +DL+ G + ++Q + +YI
Sbjct 279 PGSLLVELKRLGWVNSLKS--DSNTIAAGFGILNVTMDLSTGGLENVDEIIQLMLNYIGM 336
Query 373 LRDHGVGHELVSTMAQQSHIDFH---TTQPSSSIMDEAARLAHNLLTYEPY-HVVAGDSL 428
L+ G + +A S + F QP ++ AA L Y P H+++ L
Sbjct 337 LKSFGPQQWIHDELADLSDVKFRFKDKEQPMKMAINIAASLQ-----YIPIEHILSSRYL 391
Query 429 LIDADPRLTNQLLQEMSPSKAIIAFSDPDFTSKVDSFETDPYYGVQFRVLDL 480
L +P +LL ++PS ++ F + + +P YG + +V D+
Sbjct 392 LTKYEPERIKELLSTLTPSNMLVRVVSQKFKEQ-EGNTNEPVYGTEMKVTDI 442
> CE01818
Length=745
Score = 191 bits (485), Expect = 3e-48, Method: Compositional matrix adjust.
Identities = 130/413 (31%), Positives = 200/413 (48%), Gaps = 43/413 (10%)
Query 18 NDILKPEADYRDFRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLE 77
+ I+K D + +R +L+NG+ + V ++ + A+ G L DP ++PGLAHF E
Sbjct 15 DSIVKGSQDTKKYRGLELTNGLRVLLVSDSKTRVSAVALDVKVGHLMDPWELPGLAHFCE 74
Query 78 HMLFLGTSKYPEPESYDGFLTQNGGANNAYTDEEKTVFFNKVTDSAFEEALDRFAEFFKS 137
HMLFLGT+KYP Y +L N G +NAYTD + T + +V ALDRFA+FF
Sbjct 75 HMLFLGTAKYPSEREYFKYLAANNGDSNAYTDTDHTNYSFEVRSEKLYGALDRFAQFFLD 134
Query 138 PLFSRQYEEKEVNAIDAEHQKNIPNDDERAWYTIRSLAK--GPMSRFATGNSETLSTNPK 195
P F+ E+EV A++ E+ + D R RSL+K S+FA GN +TL +P+
Sbjct 135 PQFTESATEREVCAVNCEYLDKVNEDFWRCLQVERSLSKPGHDYSKFAIGNKKTLLEDPR 194
Query 196 AKGIDLVDRLRDFHSKYYCGSNMVAVTISPRSLDEQESLIREKLEGISAGHADWLGMVQC 255
KGI+ D L DF+ +Y M + SLD ES +LG +
Sbjct 195 TKGIEPRDVLLDFYKNWYSSDIMTCCIVGKESLDVLES---------------YLGSFK- 238
Query 256 PGPMFDTVK------------PFDHTNSGKFIHMQSFSSEPSLWVAFGLPATLTSYKKQP 303
FD +K PF K I + + + + F P + QP
Sbjct 239 ----FDAIKNTRKERKIWKDSPFGPDQLAKRIEIVPIQNTGQVSIKFPFPDLNGEFLSQP 294
Query 304 TSVLTYLLEYTGEGSLAKRLRLLGLADGISPAVDRNSISTLLG---IKVDLTQKGAAHRG 360
+ +L+ + G GSL L+ LG IS D ++I++ G + +DL+ +G H
Sbjct 295 GDYIAHLIGHEGPGSLLSELKRLGWV--ISLEADNHTIASGFGVFSVTMDLSTEGLEHVD 352
Query 361 LVLQEIFSYINFLRDHGVGHELVSTMAQQSHIDFHTTQPSSSIMDEA--ARLA 411
V+Q +F++I FL+ G + +A+ + +DF +I++E ARLA
Sbjct 353 DVIQLVFNFIGFLKSSGPQKWIHDELAELNAVDFRF--DDDTIIEETYNARLA 403
> ECU06g0750
Length=882
Score = 164 bits (414), Expect = 5e-40, Method: Compositional matrix adjust.
Identities = 105/345 (30%), Positives = 173/345 (50%), Gaps = 12/345 (3%)
Query 30 FRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYPE 89
+ + ++ NGM A+ + P ++ AV+ GS DP D GLAHFLEHMLF+GT KYP
Sbjct 27 YEYVEIPNGMRALIMSDPGLDKCSCAVSVRVGSFDDPADAQGLAHFLEHMLFMGTEKYPV 86
Query 90 PESYDGFLTQNGGANNAYTDEEKTVFFNKVTDSAFEEALDRFAEFFKSPLFSRQYEEKEV 149
+ FL++N G NA T E TV++ V AFEEA+D FA+FFKSPL R E+EV
Sbjct 87 EDGLSYFLSKNNGGYNATTYGEATVYYFDVRPEAFEEAVDMFADFFKSPLLKRDSVEREV 146
Query 150 NAIDAEHQKNIPNDDERAWYTIRSLAKG--PMSRFATGNSETLSTNPKAKGIDLVDRLRD 207
+A+++E + ND R W ++ K +S+F+TGN +TL + + + +++
Sbjct 147 SAVNSEFCNGLNNDGWRTWRMMKKCCKKEQALSKFSTGNYDTLRRDG------IWEEMKE 200
Query 208 FHSKYYCGSNMVAVTISPRSLDEQESLIREKLEGISAGHA-DWLGMVQCPGPMFDTVKPF 266
F S+ Y M V +S E + L+ K EG+ GH D + + F
Sbjct 201 FWSRKYSSDKMCVVIYGNKSPGELKELL-GKFEGVPKGHGEDSCDKKDRKCRLDEGWSVF 259
Query 267 DHTNSGKFIHMQSFSSEPSLWVAFGLPATLTSYKKQPTS-VLTYLLEYTGEGSLAKRLRL 325
D + ++I +Q + S+ V + + +K P VL +L +G A +++
Sbjct 260 DEEYTNRWIRIQPIADTRSIMVMMTVESGYKMFKNNPYEYVLNMILRNDSKG-FACKVKD 318
Query 326 LGLADGISPAVDRNSISTLLGIKVDLTQKGAAHRGLVLQEIFSYI 370
+GL + + + +++ I + L+ G + E+ Y+
Sbjct 319 MGLVLEVEGDLCDYTDYSVMIITMHLSAAGNKRPKEAVLELVKYL 363
> 7292708
Length=1077
Score = 163 bits (413), Expect = 6e-40, Method: Compositional matrix adjust.
Identities = 127/445 (28%), Positives = 212/445 (47%), Gaps = 28/445 (6%)
Query 52 AGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYPEPESYDGFLTQNGGANNAYTDEE 111
A A+ + GS +P GLAHFLEHM+F+G+ KYP+ +D + + GG NA TD E
Sbjct 94 AACALLIDYGSFAEPTKYQGLAHFLEHMIFMGSEKYPKENIFDAHIKKCGGFANANTDCE 153
Query 112 KTVFFNKVTDSAFEEALDRFAEFFKSPLFSRQYEEKEVNAIDAEHQKNIPNDDERAWYTI 171
T+F+ +V + + +LD F K+PL ++ ++E +A+D+E Q+ + +D+ R +
Sbjct 154 DTLFYFEVAEKHLDSSLDYFTALMKAPLMKQEAMQRERSAVDSEFQQILQDDETRRDQLL 213
Query 172 RSLA-KG-PMSRFATGNSETLSTNPKAKGIDLVDRLRDFHSKYYCGSNMVAVTISPR-SL 228
SLA KG P FA GN ++L N +L L + ++Y G+N + V + R +
Sbjct 214 ASLATKGFPHGTFAWGNMKSLKEN--VDDAELHKILHEIRKEHY-GANRMYVCLQARLPI 270
Query 229 DEQESLIREKLEGISAGHADWLGMVQCPG-PMFDTVKPFDHTNSGKFIHMQSFSSEPSLW 287
DE ESL+ GI V+ P F+ F + ++ +E L
Sbjct 271 DELESLVVRHFSGIPHNE------VKAPDLSSFNYKDAFKAEFHEQVFFVKPVENETKLE 324
Query 288 VAFGLPATLTSYKKQPTSVLTYLLEYTGEGSLA----KRLRLLGLADGISP-AVDRNSIS 342
+ + LP Y+ +P L+YLL Y G GSL +RL L L GI D NS+
Sbjct 325 LTWVLPNVRQYYRSKPDQFLSYLLGYEGRGSLCAYLRRRLWALQLIAGIDENGFDMNSMY 384
Query 343 TLLGIKVDLTQKGAAHRGLVLQEIFSYINFLRDHGVGHELVSTMA--QQSHIDFHTTQPS 400
+L I + LT +G + VL F+Y+ + G ++ +++ FH +P+
Sbjct 385 SLFNICIYLTDEGFKNLDEVLAATFAYVKLFANCGSMKDVYEEQQRNEETGFRFHAQRPA 444
Query 401 SSIMDEAARLAHNLLTYEPYHVVAGDSLLIDADPRLTNQL---LQEMSPSKAIIAFSDPD 457
D L NL + P ++ G L + + +L L EM + + + D
Sbjct 445 ---FDNVQELVLNLKYFPPKDILTGKELYYEYNEEHLKELISHLNEMKFNLMVTSRRKYD 501
Query 458 FTSKVDSFETDPYYGVQFRVLDLPQ 482
S D +T+ ++G ++ + +P+
Sbjct 502 DISAYD--KTEEWFGTEYATIPMPE 524
> YPR122w
Length=1208
Score = 130 bits (327), Expect = 7e-30, Method: Compositional matrix adjust.
Identities = 77/229 (33%), Positives = 119/229 (51%), Gaps = 9/229 (3%)
Query 28 RDFRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLF-LGTSK 86
R + +L NG+ A+ + P + ++ TGS DP+D+ GLAH EHM+ G+ K
Sbjct 22 RTHKVCKLPNGILALIISDPTDTSSSCSLTVCTGSHNDPKDIAGLAHLCEHMILSAGSKK 81
Query 87 YPEPESYDGFLTQNGGANNAYTDEEKTVFFNKVTDS------AFEEALDRFAEFFKSPLF 140
YP+P + + +N G+ NA+T E+T F+ ++ ++ FE LD FA FFK PLF
Sbjct 82 YPDPGLFHTLIAKNNGSQNAFTTGEQTTFYFELPNTQNNGEFTFESILDVFASFFKEPLF 141
Query 141 SRQYEEKEVNAIDAEHQKNIPNDDERAWYTIRSLAKG--PMSRFATGNSETLSTNPKAKG 198
+ KE+ AI +EH+ NI + + ++ R LA P SRF+TGN +LS+ P+ K
Sbjct 142 NPLLISKEIYAIQSEHEGNISSTTKIFYHAARILANPDHPFSRFSTGNIHSLSSIPQLKK 201
Query 199 IDLVDRLRDFHSKYYCGSNMVAVTISPRSLDEQESLIREKLEGISAGHA 247
I L L + + G N+ P+S++ L K I A
Sbjct 202 IKLKSSLNTYFENNFFGENITLCIRGPQSVNILTKLALSKFGDIKPKSA 250
> 7296373
Length=908
Score = 122 bits (306), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 104/384 (27%), Positives = 174/384 (45%), Gaps = 42/384 (10%)
Query 22 KPEADYRDFRHYQLSNGMHAIAVH---------HPRSNE-------------AGFAVAAN 59
K + D + +R LSNG+ A+ + H S E A AV
Sbjct 42 KSDGDSKLYRALTLSNGLRAMLISDSYIDEPSIHRASRESLNSSTENFNGKLAACAVLVG 101
Query 60 TGSLYDPEDVPGLAHFLEHMLFLGTSKYPEPESYDGFLTQNGGANNAYTDEEKTVFFNKV 119
GS +P+ GLAHF+EHM+F+G+ K+P +D F+T++GG +NA+T+ E+T F+ ++
Sbjct 102 VGSFSEPQQYQGLAHFVEHMIFMGSEKFPVENEFDSFVTKSGGFSNAHTENEETCFYFEL 161
Query 120 TDSAFEEALDRFAEFFKSPLFSRQYEEKEVNAIDAEHQKNIPNDDERAWYTIRSLAKG-- 177
+ + +D F K+PL +E +A+ +E ++ D+ R + SLA
Sbjct 162 DQTHLDRGMDLFMNLMKAPLMLPDAMSRERSAVQSEFEQTHMRDEVRRDQILASLASEGY 221
Query 178 PMSRFATGNSETLSTNPKAKGID---LVDRLRDFHSKYYCGSNMVAVTISPRSLDEQESL 234
P F+ GN +TL +G+D L L F+ +Y + MV + SLDE E L
Sbjct 222 PHGTFSWGNYKTLK-----EGVDDSSLHKELHKFYRDHYGSNRMVVALQAQLSLDELEEL 276
Query 235 IREKLEGISAGHADWLGMVQCPGPMFDTVKPFDHTNSGKFIHMQSFSSEPSLWVAFGLPA 294
+ I + + + Q + K F +Q L + + LP
Sbjct 277 LVRHCADIPTSQQNSIDVSQ-----LNYQKAFRDQFYKDVFLVQPVEDVCKLELTWVLPP 331
Query 295 TLTSYKKQPTSVLTYLLEYTGEGSLAKRLR----LLGLADGIS-PAVDRNSISTLLGIKV 349
Y+ +P ++ L+ Y G GSL LR + + G++ + D NSI +L I +
Sbjct 332 MKNFYRSKPDMFISQLIGYEGVGSLCAYLRHHLWCISVVAGVAESSFDSNSIYSLFNICI 391
Query 350 DLTQKGAAHRGLVLQEIFSYINFL 373
L+ G H VL+ F+++ +
Sbjct 392 YLSDDGFDHIDEVLEATFAWVKLI 415
> At3g02090
Length=531
Score = 70.9 bits (172), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 55/191 (28%), Positives = 88/191 (46%), Gaps = 10/191 (5%)
Query 31 RHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYPEP 90
R L NG+ + + A V + GS ++ ++ G AHFLEHM+F GT +
Sbjct 98 RVTTLPNGLRVATESNLSAKTATVGVWIDAGSRFESDETNGTAHFLEHMIFKGTDRR-TV 156
Query 91 ESYDGFLTQNGGANNAYTDEEKTVFFNKVTDSAFEEALDRFAEFFKSPLFSRQYEEKEVN 150
+ + + GG NAYT E+T ++ KV DS +ALD A+ ++ F Q +E +
Sbjct 157 RALEEEIEDIGGHLNAYTSREQTTYYAKVLDSNVNQALDVLADILQNSKFEEQRINRERD 216
Query 151 AIDAEHQKNIPNDDERAWYTIRSLA--KGPMSRFATGNSETLSTNPKAKGIDLVDRLRDF 208
I E Q+ DE + + A P+ R G ++ + K I D L+++
Sbjct 217 VILREMQEVEGQTDEVVLDHLHATAFQYTPLGRTILGPAQNV------KSITRED-LQNY 269
Query 209 HSKYYCGSNMV 219
+Y S MV
Sbjct 270 IKTHYTASRMV 280
> SPBP23A10.15c
Length=457
Score = 58.2 bits (139), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 50/176 (28%), Positives = 79/176 (44%), Gaps = 8/176 (4%)
Query 35 LSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYPEPESYD 94
L NG+ HHP + A V + GS + G AHFLEH+ F GT +
Sbjct 27 LKNGLTVATEHHPYAQTATVLVGVDAGSRAETAKNNGAAHFLEHLAFKGTKNRSQKALEL 86
Query 95 GFLTQNGGAN-NAYTDEEKTVFFNKVTDSAFEEALDRFAEFFKSPLFSRQYEEKEVNAID 153
F +N GA+ NAYT E+TV++ +A A+ A+ + S E+E I
Sbjct 87 EF--ENTGAHLNAYTSREQTVYYAHAFKNAVPNAVAVLADILTNSSISASAVERERQVIL 144
Query 154 AEHQKNIPNDDERAWYTIRSLA-KG-PMSRFATGNSETLSTNPKAKGIDLVDRLRD 207
E ++ DE + + + A +G P+ R G E + + + DL+ ++D
Sbjct 145 REQEEVDKMADEVVFDHLHATAYQGHPLGRTILGPKENIESLTRE---DLLQYIKD 197
> At5g42390
Length=1265
Score = 58.2 bits (139), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 64/223 (28%), Positives = 91/223 (40%), Gaps = 38/223 (17%)
Query 19 DILKPEADYRDFRHY--------------QLSNGMHAIAVHH---PRSNEAGFAVAANTG 61
D+L PE D + + QL NG+ + + + P EA V + G
Sbjct 170 DLLPPEIDSAELEAFLGCELPSHPKLHRGQLKNGLRYLILPNKVPPNRFEAHMEV--HVG 227
Query 62 SLYDPEDVPGLAHFLEHMLFLGTSKYPEPESYDGFLTQNGGANNAYTDEEKTVFF----- 116
S+ + ED G+AH +EH+ FLG+ K + L G +NAYTD TVF
Sbjct 228 SIDEEEDEQGIAHMIEHVAFLGSKKREK-------LLGTGARSNAYTDFHHTVFHIHSPT 280
Query 117 --NKVTDSAFEEALDRFAEFFKSPLFSRQYEEKEVNAIDAEHQKNIPNDDERAWYTIRSL 174
D F LD E P F EKE AI +E Q + N E + L
Sbjct 281 HTKDSEDDLFPSVLDALNEIAFHPKFLSSRVEKERRAILSELQ--MMNTIE--YRVDCQL 336
Query 175 AKGPMSRFATGNSETLSTNPKAKGIDLVDRLRDFHSKYYCGSN 217
+ S G + + K D VD++R FH ++Y +N
Sbjct 337 LQHLHSENKLGRRFPIGLEEQIKKWD-VDKIRKFHERWYFPAN 378
> At5g56730
Length=956
Score = 55.1 bits (131), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 54/221 (24%), Positives = 96/221 (43%), Gaps = 29/221 (13%)
Query 16 LGNDILKPEADYRDFRHYQLSNGMHAIAVHHPRSN-----EAGFAVAANTGSLYDPEDVP 70
LGN++ ADY +L NG+ +++ R N A A+A GS+ + ED
Sbjct 31 LGNELEPFGADYG-----RLDNGL----IYYVRRNSKPRMRAALALAVKVGSVLEEEDQR 81
Query 71 GLAHFLEHMLFLGTSKYPE---PESYDGFLTQNGGANNAYTDEEKTVF--FNKVTD-SAF 124
G+AH +EH+ F T++Y + + + G NA T ++T++ F V
Sbjct 82 GVAHIVEHLAFSATTRYTNHDIVKFLESIGAEFGPCQNAMTTADETIYELFVPVDKPELL 141
Query 125 EEALDRFAEFFKSPLFSRQYEEKEVNAIDAEHQ--KNIPNDDERAWYTIRSLAKGPMSRF 182
+A+ AEF S++ EKE A+ E++ +N + + + + R
Sbjct 142 SQAISILAEFSSEIRVSKEDLEKERGAVMEEYRGNRNATGRMQDSHWQLMMEGSKYAERL 201
Query 183 ATGNSETLSTNPKAKGIDLVDRLRDFHSKYYCGSNMVAVTI 223
G + + + P A ++ F+ K+Y NM V +
Sbjct 202 PIGLEKVIRSVPAAT-------VKQFYQKWYHLCNMAVVAV 235
> YLR163c
Length=462
Score = 54.7 bits (130), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 48/191 (25%), Positives = 81/191 (42%), Gaps = 10/191 (5%)
Query 31 RHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYPEP 90
R +L NG+ + P ++ A + + GS + G AHFLEH+ F GT +
Sbjct 27 RTSKLPNGLTIATEYIPNTSSATVGIFVDAGSRAENVKNNGTAHFLEHLAFKGTQNRSQ- 85
Query 91 ESYDGFLTQNGGANNAYTDEEKTVFFNKVTDSAFEEALDRFAEFFKSPLFSRQYEEKEVN 150
+ + + G NAYT E TV++ K +A+D ++ + E+E +
Sbjct 86 QGIELEIENIGSHLNAYTSRENTVYYAKSLQEDIPKAVDILSDILTKSVLDNSAIERERD 145
Query 151 AIDAEHQKNIPNDDERAWYTIRSLA--KGPMSRFATGNSETLSTNPKAKGIDLVDRLRDF 208
I E ++ DE + + + P+ R G + + K I D L+D+
Sbjct 146 VIIRESEEVDKMYDEVVFDHLHEITYKDQPLGRTILGPIKNI------KSITRTD-LKDY 198
Query 209 HSKYYCGSNMV 219
+K Y G MV
Sbjct 199 ITKNYKGDRMV 209
> Hs4758734
Length=489
Score = 54.3 bits (129), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 50/175 (28%), Positives = 78/175 (44%), Gaps = 12/175 (6%)
Query 59 NTGSLYDPEDVPGLAHFLEHMLFLGTSKYPEPESYDGFLTQNGGAN-NAYTDEEKTVFFN 117
+ GS Y+ E G AHFLEHM F GT K + + +N GA+ NAYT E+TV++
Sbjct 86 DAGSRYENEKNNGTAHFLEHMAFKGTKKRSQLDLE--LEIENMGAHLNAYTSREQTVYYA 143
Query 118 KVTDSAFEEALDRFAEFFKSPLFSRQYEEKEVNAIDAEHQKNIPNDDERAWYTIRSLA-- 175
K A++ A+ ++ E+E I E Q+ N E + + + A
Sbjct 144 KAFSKDLPRAVEILADIIQNSTLGEAEIERERGVILREMQEVETNLQEVVFDYLHATAYQ 203
Query 176 KGPMSRFATGNSETLSTNPKAKGIDLVDRLRDFHSKYYCGSNMVAVTISPRSLDE 230
+ R G +E + + + DLV D+ + +Y G +V S DE
Sbjct 204 NTALGRTILGPTENIKSISRK---DLV----DYITTHYKGPRIVLAAAGGVSHDE 251
> CE03806
Length=485
Score = 49.3 bits (116), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 40/122 (32%), Positives = 55/122 (45%), Gaps = 16/122 (13%)
Query 35 LSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYP----EP 90
L NG +A + + A V + GS Y+ E G AHFLEHM F GT + E
Sbjct 62 LPNGFR-VATENTGGSTATIGVFIDAGSRYENEKNNGTAHFLEHMAFKGTPRRTRMGLEL 120
Query 91 ESYDGFLTQNGGAN-NAYTDEEKTVFFNKVTDSAFEEALDRFAEFFKSPLFSRQYEEKEV 149
E +N GA+ NAYT E T ++ K F E LD+ + L + K++
Sbjct 121 E------VENIGAHLNAYTSRESTTYYAK----CFTEKLDQSVDILSDILLNSSLATKDI 170
Query 150 NA 151
A
Sbjct 171 EA 172
> YOL098c
Length=1037
Score = 46.2 bits (108), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 45/181 (24%), Positives = 75/181 (41%), Gaps = 20/181 (11%)
Query 71 GLAHFLEHMLFLGTSKYPEPESYDGFLTQNGGAN----NAYTDEEKTVF-FNKVTDSAFE 125
G H LEH++F+G+ YP Y G L G + NA+TD ++TV+ + F
Sbjct 57 GAPHTLEHLIFMGSKSYP----YKGLLDTAGNLSLSNTNAWTDTDQTVYTLSSAGWKGFS 112
Query 126 EALDRFAEFFKSPLFSRQYEEKEVNAIDAEHQKN-------IPNDDERAWYTIRSLAKGP 178
+ L + + P + + EV ID E+ + + + + WY I L K
Sbjct 113 KLLPAYLDHILHPTLTDEACLTEVYHIDPENLGDKGVVFSEMEAIETQGWY-ISGLEKQR 171
Query 179 MSRFATGNSETLSTNPKAKGIDLV--DRLRDFHSKYYCGSNMVAVTISPRSLDEQESLIR 236
+ F G+ T K + + D +R FH Y N+ + DE +++
Sbjct 172 L-MFPEGSGYRSETGGLTKNLRTLTNDEIRQFHKSLYSSDNLCVIVCGNVPTDELLTVME 230
Query 237 E 237
E
Sbjct 231 E 231
> Hs4507841
Length=480
Score = 45.4 bits (106), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 48/216 (22%), Positives = 87/216 (40%), Gaps = 11/216 (5%)
Query 35 LSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYPEPESYD 94
L NG+ +A V + GS ++ E G +FLEH+ F GT P + +
Sbjct 53 LDNGLR-VASEQSSQPTCTVGVWIDVGSRFETEKNNGAGYFLEHLAFKGTKNRP-GSALE 110
Query 95 GFLTQNGGANNAYTDEEKTVFFNKVTDSAFEEALDRFAEFFKSPLFSRQYEEKEVNAIDA 154
+ G NAY+ E T ++ K +A++ + ++ EKE + I
Sbjct 111 KEVESMGAHLNAYSTREHTAYYIKALSKDLPKAVELLGDIVQNCSLEDSQIEKERDVILR 170
Query 155 EHQKNIPNDDERAWYTIRSLA--KGPMSRFATGNSETLSTNPKAKGIDLVDRLRDFHSKY 212
E Q+N + + + + + A P+++ G SE + +A L ++ S +
Sbjct 171 EMQENDASMRDVVFNYLHATAFQGTPLAQAVEGPSENVRKLSRAD-------LTEYLSTH 223
Query 213 YCGSNMVAVTISPRSLDEQESLIREKLEGISAGHAD 248
Y MV + L ++ L GI +A+
Sbjct 224 YKAPRMVLAAAGGVEHQQLLDLAQKHLGGIPWTYAE 259
> CE03926
Length=995
Score = 45.1 bits (105), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 41/169 (24%), Positives = 73/169 (43%), Gaps = 22/169 (13%)
Query 71 GLAHFLEHMLFLGTSKYPEPESYDGFLTQN-GGANNAYTDEEKTVF-FNKVTDSAFEEAL 128
GL H LEH++F+G+ KYP D + NA+TD + T + + V F + L
Sbjct 57 GLPHTLEHLVFMGSKKYPFKGVLDVIANRCLADGTNAWTDTDHTAYTLSTVGSDGFLKVL 116
Query 129 DRFAEFFKSPLFSRQYEEKEVNAIDAE-------------HQKNIPNDDERAWYTIRSLA 175
+ +P+ + EV+ I E H+ + + +R + +
Sbjct 117 PVYINHLLTPMLTASQFATEVHHITGEGNDAGVVYSEMQDHESEMESIMDR---KTKEVI 173
Query 176 KGPMSRFATGNSETLSTNPKAKGIDLVDRLRDFHSKYYCGSNMVAVTIS 224
P + +A L ++ + +++RD+H K+Y SNMV VT+
Sbjct 174 YPPFNPYAVDTGGRLKNLRESCTL---EKVRDYHKKFYHLSNMV-VTVC 218
> SPAC3H1.02c
Length=1036
Score = 44.3 bits (103), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 45/200 (22%), Positives = 79/200 (39%), Gaps = 24/200 (12%)
Query 36 SNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYPEPESYDG 95
+ G +V P S G V A + D G H LEH+ F+G+ KYP
Sbjct 28 ATGFSVASVKTPTSRLQGSFVVAT-----EAHDNLGCPHTLEHLCFMGSKKYPMNGILTK 82
Query 96 FLTQNGGANNAYTDEEKTVF-FNKVTDSAFEEALDRFAEFFKSPLFSRQYEEKEVNAIDA 154
F + G NA TD + T + + + F L FA+ SP+ S + EV I+
Sbjct 83 FAGRACGDINACTDVDYTSYELSAAEEDGFLRLLPVFADHILSPILSDEAFCTEVYHING 142
Query 155 ---------EHQKNIPNDDERAWYTIRSLAKGPMSR---FATGNSETLSTNPKAKGIDLV 202
+N + + + ++ P++ + TG +P +
Sbjct 143 MGEESGVVYSEMQNTQSSETDVMFDCMRTSQYPVTSGYYYETGG------HPSELRKLSI 196
Query 203 DRLRDFHSKYYCGSNMVAVT 222
+++R++H + Y SN+ +
Sbjct 197 EKIREYHKEMYVPSNICLIV 216
> CE11226
Length=471
Score = 41.6 bits (96), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 32/117 (27%), Positives = 54/117 (46%), Gaps = 3/117 (2%)
Query 35 LSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYPEPESYD 94
L NG + + S A V TGS ++ E G+AHFLE ++ GT K + +
Sbjct 43 LKNGFRVVTEDNG-SATATVGVWIETGSRFENEKNNGVAHFLERLIHKGTGKRASA-ALE 100
Query 95 GFLTQNGGANNAYTDEEKTVFFNKVTDSAFEEALDRFAEFFK-SPLFSRQYEEKEVN 150
L G N++T+ ++T F + E+ +D A+ + S L + + + VN
Sbjct 101 SELNAIGAKLNSFTERDQTAVFVQAGAQDVEKVVDILADVLRNSKLEASTIDTERVN 157
> Hs15321446
Length=525
Score = 40.8 bits (94), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 26/121 (21%), Positives = 52/121 (42%), Gaps = 1/121 (0%)
Query 35 LSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYPEPESYD 94
L NG+ +A + + N+GS Y+ + + G+AHFLE + F T+++ +
Sbjct 72 LDNGLR-VASQNKFGQFCTVGILINSGSRYEAKYLSGIAHFLEKLAFSSTARFDSKDEIL 130
Query 95 GFLTQNGGANNAYTDEEKTVFFNKVTDSAFEEALDRFAEFFKSPLFSRQYEEKEVNAIDA 154
L ++GG + T + T++ + + A+ P + + E A+
Sbjct 131 LTLEKHGGICDCQTSRDTTMYAVSADSKGLDTVVALLADVVLQPRLTDEEVEMTRMAVQF 190
Query 155 E 155
E
Sbjct 191 E 191
> 7304131
Length=556
Score = 37.0 bits (84), Expect = 0.097, Method: Compositional matrix adjust.
Identities = 26/122 (21%), Positives = 50/122 (40%), Gaps = 3/122 (2%)
Query 35 LSNGMHAIAVHHPRSNE-AGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYPEPESY 93
L NG+ + PR + + ++G Y+ G++HFLE + F T +P ++
Sbjct 99 LPNGLRIAS--EPRYGQFCTVGLVIDSGPRYEVAYPSGVSHFLEKLAFNSTVNFPNKDAI 156
Query 94 DGFLTQNGGANNAYTDEEKTVFFNKVTDSAFEEALDRFAEFFKSPLFSRQYEEKEVNAID 153
L +NGG + + + ++ + A + A+ P S Q A++
Sbjct 157 LKELEKNGGICDCQSSRDTLIYAASIDSRAIDSVTRLLADVTLRPTLSDQEVSLARRAVN 216
Query 154 AE 155
E
Sbjct 217 FE 218
> YBR097w
Length=1454
Score = 36.2 bits (82), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 33/111 (29%), Positives = 51/111 (45%), Gaps = 16/111 (14%)
Query 57 AANTGSLYDPEDVPGLAHFLEHMLFLGTSK----YPEPESYDGFLTQNGGANNAYTDEEK 112
AA +Y PED PG E + + TSK Y PE ++ L Q+G +NN +E
Sbjct 167 AAFIKPVYLPEDNPG-----EFLFYFDTSKRRTCYLAPERFNSKLYQDGKSNNGRLTKEM 221
Query 113 TVF-FNKVTDSAFEEALDRF--AEFFK----SPLFSRQYEEKEVNAIDAEH 156
+F V F E F ++ FK S +R++ +E+N+ D +
Sbjct 222 DIFSLGCVIAEIFAEGRPIFNLSQLFKYKSNSYDVNREFLMEEMNSTDLRN 272
> Hs20535969
Length=417
Score = 35.8 bits (81), Expect = 0.25, Method: Compositional matrix adjust.
Identities = 24/103 (23%), Positives = 44/103 (42%), Gaps = 4/103 (3%)
Query 55 AVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYPEPESYDGFLTQNGGANNAYTDEEKTV 114
+ N+GS Y+ + G+AHFLE + F +++ + L ++GG + T + T
Sbjct 39 GILINSGSRYEATYLSGIAHFLEKLAFSSNARFNSKDEILLTLEKHGGICDCQTSSDTTT 98
Query 115 FFNKVTDSAFEEALDRFAEFFKSPLFSRQYEEKEVNAIDAEHQ 157
+ SA + LD + Q ++EV + Q
Sbjct 99 Y----AVSADSKGLDTVFGLLADAVLQPQLTDEEVEMMQMAVQ 137
> At3g19170
Length=1052
Score = 34.3 bits (77), Expect = 0.60, Method: Compositional matrix adjust.
Identities = 26/89 (29%), Positives = 42/89 (47%), Gaps = 12/89 (13%)
Query 30 FRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYPE 89
F+H + G ++V + N+ F V T P+D G+ H LEH + G+ KYP
Sbjct 97 FKHKK--TGCEVMSVSNEDENKV-FGVVFRT----PPKDSTGIPHILEHSVLCGSRKYPV 149
Query 90 PESYDGFLTQNGGAN---NAYTDEEKTVF 115
E + L G + NA+T ++T +
Sbjct 150 KEPFVELL--KGSLHTFLNAFTYPDRTCY 176
> At1g49630
Length=1076
Score = 34.3 bits (77), Expect = 0.70, Method: Compositional matrix adjust.
Identities = 25/89 (28%), Positives = 42/89 (47%), Gaps = 12/89 (13%)
Query 30 FRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYPE 89
F+H + G ++V + N+ F + T P+D G+ H LEH + G+ KYP
Sbjct 124 FKHKK--TGCEVMSVSNDDENKV-FGIVFRT----PPKDSTGIPHILEHSVLCGSRKYPM 176
Query 90 PESYDGFLTQNGGAN---NAYTDEEKTVF 115
E + L G + NA+T ++T +
Sbjct 177 KEPFVELL--KGSLHTFLNAFTYPDRTCY 203
> 7299936
Length=382
Score = 33.1 bits (74), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 40/169 (23%), Positives = 70/169 (41%), Gaps = 12/169 (7%)
Query 79 MLFLGTSKYPEPESYDGFLTQNGGAN-NAYTDEEKTVFFNKVTDSAFEEALDRFAEFFKS 137
M F GT+K + + +N GA+ NAYT E+TVF+ K +A++ A+ ++
Sbjct 1 MAFKGTAKRSQTDLE--LEVENLGAHLNAYTSREQTVFYAKCLSKDVPKAVEILADIIQN 58
Query 138 PLFSRQYEEKEVNAIDAEHQKNIPNDDERAWYTIRSLA--KGPMSRFATGNSETLSTNPK 195
+E + I E Q+ N E + + + A P+ + G ++ + + K
Sbjct 59 SKLGEAEIARERSVILREMQEVESNLQEVVFDHLHATAYQGTPLGQTILGPTKNIQSIGK 118
Query 196 AKGIDLVDRLRDFHSKYYCGSNMVAVTISPRSLDEQESLIREKLEGISA 244
A L D+ +Y S +V D+ L L G+ A
Sbjct 119 AD-------LTDYIQTHYKASRIVLAAAGGVKHDDLVKLACSSLGGLEA 160
> At5g40740
Length=752
Score = 32.3 bits (72), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 62/224 (27%), Positives = 96/224 (42%), Gaps = 46/224 (20%)
Query 275 IHMQSFSSEPSLWVAFGLPATLT--SYKKQPTSVLTYLLEYTGEGSLAKRLRLLGLADGI 332
+H ++F P+ + LP++LT S+ T LL T + +R R L A+
Sbjct 175 VHRRTF---PADVASNPLPSSLTDVSFSHAAT-----LLPVTKARIVLERRRFLKNAE-- 224
Query 333 SPAVDRNSISTLLGIKVDLTQKGAAHRGLVLQEIFSYINFLRDH-----GVGHELVSTMA 387
AV R ++ + L ++ +G LQ+ +N LR+ V +LVS+ +
Sbjct 225 -TAVQRQAMWSNLAHEMTAEFRGLCAEEAYLQQELEKLNDLRNKVKQEGEVWDDLVSSSS 283
Query 388 QQSHIDFHTTQPSSSIMDEAARLAHNLLTYEPY----------HVVAGDSLLIDAD---- 433
Q SH+ T+ SIM A + H +L P + ++G +LL D
Sbjct 284 QNSHLVSKATRLWDSIM--ARKGQHEVLASGPIEDLIAHREHRYRISGSALLAAMDQSSQ 341
Query 434 -PRLTNQLLQEMSPSKAIIA----FSDPDFT-----SKVDSFET 467
PR +LL S A +A SD +T S VDSFET
Sbjct 342 VPRA--ELLSAHSDDSASLADDKELSDGSYTNMHDHSLVDSFET 383
> Hs7657343
Length=1038
Score = 31.6 bits (70), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 31/134 (23%), Positives = 51/134 (38%), Gaps = 25/134 (18%)
Query 32 HYQLSNGMHAIAVHHPRSNEAGFAVAA-----NTGSLY------------------DPED 68
Y+L + +H V+ S F A +TG+ Y P D
Sbjct 39 QYKLGDKIHGFTVNQVTSVPELFLTAVKLTHDDTGARYLHLAREDTNNLFSVQFRTTPMD 98
Query 69 VPGLAHFLEHMLFLGTSKYPEPESYDGFLTQNGGA-NNAYTDEEKTVF-FNKVTDSAFEE 126
G+ H LEH + G+ KYP + L ++ NA+T + T++ F+ F+
Sbjct 99 STGVPHILEHTVLCGSQKYPCRNPFFKMLNRSLSTFMNAFTASDYTLYPFSTQNPKDFQN 158
Query 127 ALDRFAEFFKSPLF 140
L + + SP
Sbjct 159 LLSVYLDATFSPCL 172
> 7302255
Length=1112
Score = 31.2 bits (69), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 17/51 (33%), Positives = 26/51 (50%), Gaps = 1/51 (1%)
Query 66 PEDVPGLAHFLEHMLFLGTSKYPEPESYDGFLTQNGGA-NNAYTDEEKTVF 115
P D GL H LEH+ G+ KYP + + L ++ NA T + T++
Sbjct 120 PFDSTGLPHILEHLSLCGSQKYPVRDPFFKMLNRSVATFMNAMTGPDYTIY 170
> At3g50310
Length=342
Score = 30.8 bits (68), Expect = 6.5, Method: Compositional matrix adjust.
Identities = 29/97 (29%), Positives = 44/97 (45%), Gaps = 7/97 (7%)
Query 279 SFSSEPSLWVAFG-LPATLTSYKKQPT-----SVLTYLLEYTGEGSLAKRLRLLGLADGI 332
S S+E S+ + G P + Y + T + LLEY GSLA ++ LG
Sbjct 45 SLSNEKSVLDSLGDCPEIIRCYGEDSTVENGEEMHNLLLEYASRGSLASYMKKLGGEGLP 104
Query 333 SPAVDRNSISTLLGIKVDLTQKGAAHRGLVLQEIFSY 369
V R++ S L G++ + KG AH + L I +
Sbjct 105 ESTVRRHTGSVLRGLR-HIHAKGFAHCDIKLANILLF 140
> YDR430c
Length=989
Score = 30.8 bits (68), Expect = 7.2, Method: Compositional matrix adjust.
Identities = 20/63 (31%), Positives = 30/63 (47%), Gaps = 5/63 (7%)
Query 54 FAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYPEPESYDGFLTQN-GGANNAYTDEEK 112
F++A T +P D G+ H LEH G+ KYP + + L ++ NA T +
Sbjct 68 FSIAFKT----NPPDSTGVPHILEHTTLCGSVKYPVRDPFFKMLNKSLANFMNAMTGPDY 123
Query 113 TVF 115
T F
Sbjct 124 TFF 126
Lambda K H
0.317 0.134 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 12965481912
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40