bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_1292_orf1
Length=90
Score E
Sequences producing significant alignments: (Bits) Value
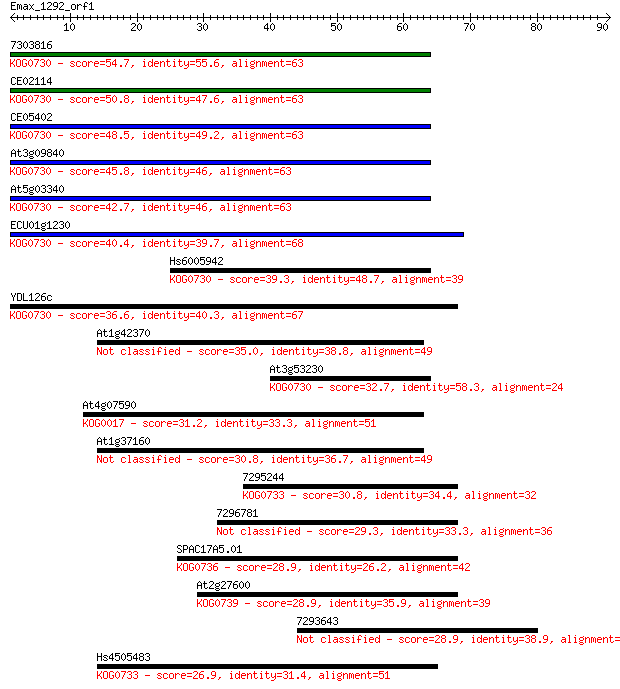
7303816 54.7 4e-08
CE02114 50.8 6e-07
CE05402 48.5 3e-06
At3g09840 45.8 2e-05
At5g03340 42.7 2e-04
ECU01g1230 40.4 8e-04
Hs6005942 39.3 0.002
YDL126c 36.6 0.013
At1g42370 35.0 0.031
At3g53230 32.7 0.15
At4g07590 31.2 0.46
At1g37160 30.8 0.62
7295244 30.8 0.67
7296781 29.3 2.0
SPAC17A5.01 28.9 2.3
At2g27600 28.9 2.7
7293643 28.9 2.7
Hs4505483 26.9 8.7
> 7303816
Length=801
Score = 54.7 bits (130), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 35/67 (52%), Positives = 39/67 (58%), Gaps = 4/67 (5%)
Query 1 RAAKAAIRDAIAAE---ELARAAG-NEGMDEDESNVKYEITRKHFEEGIAGARRSVSQTD 56
RA K AIR AI AE E RA N MD DE + EIT HFEE + ARRSVS D
Sbjct 690 RACKLAIRQAIEAEIRREKERAENQNSAMDMDEDDPVPEITSAHFEEAMKFARRSVSDND 749
Query 57 LSKYDSF 63
+ KY+ F
Sbjct 750 IRKYEMF 756
> CE02114
Length=809
Score = 50.8 bits (120), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 30/70 (42%), Positives = 41/70 (58%), Gaps = 7/70 (10%)
Query 1 RAAKAAIRDAIAAE-------ELARAAGNEGMDEDESNVKYEITRKHFEEGIAGARRSVS 53
RA K AIR++I E + +A G E M++D + EITR HFEE + ARRSV+
Sbjct 700 RACKLAIRESIEKEIRIEKERQDRQARGEELMEDDAVDPVPEITRAHFEEAMKFARRSVT 759
Query 54 QTDLSKYDSF 63
D+ KY+ F
Sbjct 760 DNDIRKYEMF 769
> CE05402
Length=810
Score = 48.5 bits (114), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 31/70 (44%), Positives = 42/70 (60%), Gaps = 7/70 (10%)
Query 1 RAAKAAIRDAIAAE---ELAR----AAGNEGMDEDESNVKYEITRKHFEEGIAGARRSVS 53
RA K AIR++I E E R A G E M+++ ++ EITR HFEE + ARRSV+
Sbjct 698 RACKLAIRESIEREIRQEKERQDRSARGEELMEDELADPVPEITRAHFEEAMKFARRSVT 757
Query 54 QTDLSKYDSF 63
D+ KY+ F
Sbjct 758 DNDIRKYEMF 767
> At3g09840
Length=809
Score = 45.8 bits (107), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 29/66 (43%), Positives = 37/66 (56%), Gaps = 3/66 (4%)
Query 1 RAAKAAIRDAIAAE---ELARAAGNEGMDEDESNVKYEITRKHFEEGIAGARRSVSQTDL 57
RA K AIR+ I + E R+ E M+ED + EI HFEE + ARRSVS D+
Sbjct 697 RACKYAIRENIEKDIEKEKRRSENPEAMEEDGVDEVSEIKAAHFEESMKYARRSVSDADI 756
Query 58 SKYDSF 63
KY +F
Sbjct 757 RKYQAF 762
> At5g03340
Length=843
Score = 42.7 bits (99), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 29/67 (43%), Positives = 38/67 (56%), Gaps = 4/67 (5%)
Query 1 RAAKAAIRDAIAAE---ELARAAGNEGMDEDESNVKY-EITRKHFEEGIAGARRSVSQTD 56
RA K AIR+ I + E R+ E M+ED + + EI HFEE + ARRSVS D
Sbjct 729 RACKYAIRENIEKDIENERRRSQNPEAMEEDMVDDEVSEIRAAHFEESMKYARRSVSDAD 788
Query 57 LSKYDSF 63
+ KY +F
Sbjct 789 IRKYQAF 795
> ECU01g1230
Length=780
Score = 40.4 bits (93), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 27/70 (38%), Positives = 39/70 (55%), Gaps = 4/70 (5%)
Query 1 RAAKAAIRDAIAAEELARAAGNEGMDEDESNVKYEITRKHFEEGIAGARRSVSQTDLSKY 60
RA K AIR+ I E + G+E MD E V Y + H + + ARRSVS+ ++ +Y
Sbjct 696 RACKLAIRETIEYELEQKKKGSEMMDL-EDPVPY-LRPDHLVQSLKTARRSVSEKEVERY 753
Query 61 DSF--RMKFD 68
++F MK D
Sbjct 754 EAFARSMKVD 763
> Hs6005942
Length=806
Score = 39.3 bits (90), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 19/39 (48%), Positives = 25/39 (64%), Gaps = 0/39 (0%)
Query 25 MDEDESNVKYEITRKHFEEGIAGARRSVSQTDLSKYDSF 63
M+ +E + EI R HFEE + ARRSVS D+ KY+ F
Sbjct 720 MEVEEDDPVPEIRRDHFEEAMRFARRSVSDNDIRKYEMF 758
> YDL126c
Length=835
Score = 36.6 bits (83), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 27/82 (32%), Positives = 41/82 (50%), Gaps = 15/82 (18%)
Query 1 RAAKAAIRDAIAAE---ELARAAGNEGMD------------EDESNVKYEITRKHFEEGI 45
RAAK AI+D+I A E + EG D E E + IT++HF E +
Sbjct 703 RAAKYAIKDSIEAHRQHEAEKEVKVEGEDVEMTDEGAKAEQEPEVDPVPYITKEHFAEAM 762
Query 46 AGARRSVSQTDLSKYDSFRMKF 67
A+RSVS +L +Y+++ +
Sbjct 763 KTAKRSVSDAELRRYEAYSQQM 784
> At1g42370
Length=619
Score = 35.0 bits (79), Expect = 0.031, Method: Composition-based stats.
Identities = 19/49 (38%), Positives = 31/49 (63%), Gaps = 1/49 (2%)
Query 14 EELARAAGNEGMDEDESNVKYEITRKHFEEGIAGARRSVSQTDLSKYDS 62
++L A ++GMDED + VK++ + HF E + AR+S S T+ K +S
Sbjct 411 DDLLEEAASDGMDEDRA-VKFDTSMYHFAEHVPPARQSKSLTEAHKNNS 458
> At3g53230
Length=815
Score = 32.7 bits (73), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 14/24 (58%), Positives = 17/24 (70%), Gaps = 0/24 (0%)
Query 40 HFEEGIAGARRSVSQTDLSKYDSF 63
HFEE + ARRSVS D+ KY +F
Sbjct 738 HFEESMKYARRSVSDADIRKYQAF 761
> At4g07590
Length=860
Score = 31.2 bits (69), Expect = 0.46, Method: Composition-based stats.
Identities = 17/51 (33%), Positives = 28/51 (54%), Gaps = 1/51 (1%)
Query 12 AAEELARAAGNEGMDEDESNVKYEITRKHFEEGIAGARRSVSQTDLSKYDS 62
A + L A +GMDED V+++ + HF E + AR S S ++ + +S
Sbjct 647 AEDVLIEEATEDGMDEDRE-VEFDTSMYHFSEHVPPARESKSMSEAHRNNS 696
> At1g37160
Length=480
Score = 30.8 bits (68), Expect = 0.62, Method: Composition-based stats.
Identities = 18/49 (36%), Positives = 30/49 (61%), Gaps = 1/49 (2%)
Query 14 EELARAAGNEGMDEDESNVKYEITRKHFEEGIAGARRSVSQTDLSKYDS 62
++L A ++GMDED + V+++ + HF E + AR S S T+ K +S
Sbjct 269 DDLLEEAASDGMDEDRA-VEFDTSVYHFGEHVPLARESKSLTEAHKNNS 316
> 7295244
Length=944
Score = 30.8 bits (68), Expect = 0.67, Method: Compositional matrix adjust.
Identities = 11/32 (34%), Positives = 19/32 (59%), Gaps = 0/32 (0%)
Query 36 ITRKHFEEGIAGARRSVSQTDLSKYDSFRMKF 67
+ +HF+E + R SV++ D YD R+K+
Sbjct 902 VRSQHFQEALQQLRPSVNEQDRKIYDKLRLKY 933
> 7296781
Length=554
Score = 29.3 bits (64), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 12/36 (33%), Positives = 21/36 (58%), Gaps = 0/36 (0%)
Query 32 VKYEITRKHFEEGIAGARRSVSQTDLSKYDSFRMKF 67
V ++ K F E ++ +SVS+ DL KY+ + +F
Sbjct 516 VDLPVSNKDFNEAMSRCNKSVSRADLDKYEKWMREF 551
> SPAC17A5.01
Length=948
Score = 28.9 bits (63), Expect = 2.3, Method: Composition-based stats.
Identities = 11/42 (26%), Positives = 26/42 (61%), Gaps = 0/42 (0%)
Query 26 DEDESNVKYEITRKHFEEGIAGARRSVSQTDLSKYDSFRMKF 67
+E++ +++ IT++ F + R S+S+ +L +Y+ R +F
Sbjct 906 NENQDSLELRITKEDFLTSLKKLRPSISEQELHRYEMVRHQF 947
> At2g27600
Length=435
Score = 28.9 bits (63), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 14/39 (35%), Positives = 23/39 (58%), Gaps = 0/39 (0%)
Query 29 ESNVKYEITRKHFEEGIAGARRSVSQTDLSKYDSFRMKF 67
E + ITR FE+ +A R +VS++DL ++ F +F
Sbjct 393 EKIIPPPITRTDFEKVLARQRPTVSKSDLDVHERFTQEF 431
> 7293643
Length=381
Score = 28.9 bits (63), Expect = 2.7, Method: Composition-based stats.
Identities = 14/36 (38%), Positives = 23/36 (63%), Gaps = 0/36 (0%)
Query 44 GIAGARRSVSQTDLSKYDSFRMKFDPIYKSQAAGGD 79
GIA RRS S+T ++ ++ + KF +Y+ + A GD
Sbjct 281 GIAPVRRSQSRTRETREEARQRKFRYLYQVRTARGD 316
> Hs4505483
Length=750
Score = 26.9 bits (58), Expect = 8.7, Method: Composition-based stats.
Identities = 16/52 (30%), Positives = 27/52 (51%), Gaps = 8/52 (15%)
Query 14 EELAR-AAGNEGMDEDESNVKYEITRKHFEEGIAGARRSVSQTDLSKYDSFR 64
+E+AR +GNE + +++ KHFEE R S+S+ D Y+ +
Sbjct 701 QEMARQKSGNE-------KGELKVSHKHFEEAFKKVRSSISKKDQIMYERLQ 745
Lambda K H
0.312 0.129 0.360
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1177758614
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40