bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
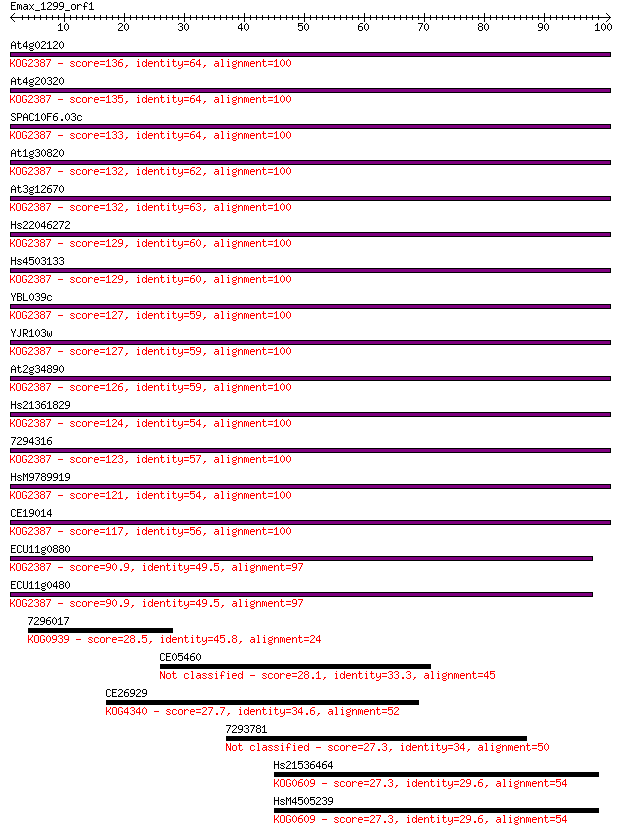
Query= Emax_1299_orf1
Length=100
Score E
Sequences producing significant alignments: (Bits) Value
At4g02120 136 1e-32
At4g20320 135 1e-32
SPAC10F6.03c 133 9e-32
At1g30820 132 2e-31
At3g12670 132 2e-31
Hs22046272 129 1e-30
Hs4503133 129 1e-30
YBL039c 127 4e-30
YJR103w 127 6e-30
At2g34890 126 1e-29
Hs21361829 124 6e-29
7294316 123 7e-29
HsM9789919 121 3e-28
CE19014 117 4e-27
ECU11g0880 90.9 6e-19
ECU11g0480 90.9 6e-19
7296017 28.5 3.0
CE05460 28.1 4.2
CE26929 27.7 5.5
7293781 27.3 7.0
Hs21536464 27.3 7.3
HsM4505239 27.3 7.4
> At4g02120
Length=539
Score = 136 bits (342), Expect = 1e-32, Method: Composition-based stats.
Identities = 64/108 (59%), Positives = 81/108 (75%), Gaps = 8/108 (7%)
Query 1 PIQHGEVFVTDDGAETDLDLGHYERFIRTKMTRRNNFTTGRVYSEVLRKERRGDYLGATI 60
P +HGEVFV DDG E DLDLG+YERF+ +T+ NN TTG++Y VL KER+GDYLG T+
Sbjct 52 PFEHGEVFVLDDGGEVDLDLGNYERFLDVTLTKDNNITTGKIYQSVLDKERKGDYLGKTV 111
Query 61 QVIPHITNEIKERI-------IRGGEGH-DVVLVEVGGTVGDIESLPF 100
QV+PHIT+ IK+ I + G EG DV ++E+GGTVGDIES+PF
Sbjct 112 QVVPHITDAIKDWIESVSLIPVDGKEGQADVCVIELGGTVGDIESMPF 159
> At4g20320
Length=553
Score = 135 bits (341), Expect = 1e-32, Method: Composition-based stats.
Identities = 64/108 (59%), Positives = 81/108 (75%), Gaps = 8/108 (7%)
Query 1 PIQHGEVFVTDDGAETDLDLGHYERFIRTKMTRRNNFTTGRVYSEVLRKERRGDYLGATI 60
PI+HGEV+V DDG E DLDLG+YERF+ K+T NN TTG+VY VL KERRGDYLG T+
Sbjct 52 PIEHGEVYVLDDGGEVDLDLGNYERFMDIKLTSENNITTGKVYKHVLEKERRGDYLGKTV 111
Query 61 QVIPHITNEIKERIIRGG----EGH----DVVLVEVGGTVGDIESLPF 100
QV+PHIT+ I++ I R +G DV ++E+GGT+GDIES+PF
Sbjct 112 QVVPHITDAIQKWIERAARIPVDGQSGPADVCVIELGGTIGDIESMPF 159
> SPAC10F6.03c
Length=600
Score = 133 bits (334), Expect = 9e-32, Method: Compositional matrix adjust.
Identities = 64/108 (59%), Positives = 82/108 (75%), Gaps = 8/108 (7%)
Query 1 PIQHGEVFVTDDGAETDLDLGHYERFIRTKMTRRNNFTTGRVYSEVLRKERRGDYLGATI 60
P++HGEVFV +DG E DLDLG+YER++ +T NN TTG+VYS V++KERRGDYLG T+
Sbjct 52 PLEHGEVFVLNDGGEVDLDLGNYERYLNVTLTHDNNITTGKVYSNVIQKERRGDYLGKTV 111
Query 61 QVIPHITNEIK---ERIIR-----GGEGHDVVLVEVGGTVGDIESLPF 100
Q++PH+TNEI+ ER+ R GE DV +VE+GGTVGDIES F
Sbjct 112 QIVPHVTNEIQDWVERVARIPVDQSGEEPDVCIVELGGTVGDIESAAF 159
> At1g30820
Length=600
Score = 132 bits (332), Expect = 2e-31, Method: Composition-based stats.
Identities = 62/108 (57%), Positives = 80/108 (74%), Gaps = 8/108 (7%)
Query 1 PIQHGEVFVTDDGAETDLDLGHYERFIRTKMTRRNNFTTGRVYSEVLRKERRGDYLGATI 60
P +HGEVFV DDG E DLDLG+YERF+ K+TR NN TTG++Y V+ KER+GDYLG T+
Sbjct 52 PFEHGEVFVLDDGGEVDLDLGNYERFLDIKLTRDNNITTGKIYQHVIAKERKGDYLGKTV 111
Query 61 QVIPHITNEIKERIIR------GGEGH--DVVLVEVGGTVGDIESLPF 100
QV+PH+T+ I++ I R GE DV ++E+GGT+GDIES PF
Sbjct 112 QVVPHVTDAIQDWIERVAVIPVDGEEDPADVCVIELGGTIGDIESAPF 159
> At3g12670
Length=591
Score = 132 bits (331), Expect = 2e-31, Method: Compositional matrix adjust.
Identities = 63/108 (58%), Positives = 82/108 (75%), Gaps = 8/108 (7%)
Query 1 PIQHGEVFVTDDGAETDLDLGHYERFIRTKMTRRNNFTTGRVYSEVLRKERRGDYLGATI 60
P +HGEVFV DDG E DLDLG+YERF+ + +TR NN TTG++Y V+ KER+GDYLG T+
Sbjct 52 PFEHGEVFVLDDGGEVDLDLGNYERFLDSTLTRDNNITTGKIYQSVIDKERKGDYLGRTV 111
Query 61 QVIPHITNEIKERIIR-------GGEG-HDVVLVEVGGTVGDIESLPF 100
QV+PH+T+ I+E I R G EG DV ++E+GGT+GDIES+PF
Sbjct 112 QVVPHVTDAIQEWIERVANVPVDGKEGPPDVCVIELGGTIGDIESMPF 159
> Hs22046272
Length=591
Score = 129 bits (324), Expect = 1e-30, Method: Compositional matrix adjust.
Identities = 60/108 (55%), Positives = 80/108 (74%), Gaps = 8/108 (7%)
Query 1 PIQHGEVFVTDDGAETDLDLGHYERFIRTKMTRRNNFTTGRVYSEVLRKERRGDYLGATI 60
P +HGEVFV DDG E DLDLG+YERF+ ++T+ NN TTG++Y V+ KER+GDYLG T+
Sbjct 52 PYEHGEVFVLDDGGEVDLDLGNYERFLDIRLTKDNNLTTGKIYQYVINKERKGDYLGKTV 111
Query 61 QVIPHITNEIKERIIR--------GGEGHDVVLVEVGGTVGDIESLPF 100
QV+PHIT+ I+E ++R G V ++E+GGTVGDIES+PF
Sbjct 112 QVVPHITDAIQEWVMRQALIPVDEDGLEPQVCVIELGGTVGDIESMPF 159
> Hs4503133
Length=591
Score = 129 bits (324), Expect = 1e-30, Method: Compositional matrix adjust.
Identities = 60/108 (55%), Positives = 80/108 (74%), Gaps = 8/108 (7%)
Query 1 PIQHGEVFVTDDGAETDLDLGHYERFIRTKMTRRNNFTTGRVYSEVLRKERRGDYLGATI 60
P +HGEVFV DDG E DLDLG+YERF+ ++T+ NN TTG++Y V+ KER+GDYLG T+
Sbjct 52 PYEHGEVFVLDDGGEVDLDLGNYERFLDIRLTKDNNLTTGKIYQYVINKERKGDYLGKTV 111
Query 61 QVIPHITNEIKERIIR--------GGEGHDVVLVEVGGTVGDIESLPF 100
QV+PHIT+ I+E ++R G V ++E+GGTVGDIES+PF
Sbjct 112 QVVPHITDAIQEWVMRQALIPVDEDGLEPQVCVIELGGTVGDIESMPF 159
> YBL039c
Length=579
Score = 127 bits (320), Expect = 4e-30, Method: Compositional matrix adjust.
Identities = 59/108 (54%), Positives = 80/108 (74%), Gaps = 8/108 (7%)
Query 1 PIQHGEVFVTDDGAETDLDLGHYERFIRTKMTRRNNFTTGRVYSEVLRKERRGDYLGATI 60
P++HGE FV DDG ETDLDLG+YER++ +T+ +N TTG++YS V+ KER+GDYLG T+
Sbjct 52 PLEHGECFVLDDGGETDLDLGNYERYLGVTLTKDHNITTGKIYSHVIAKERKGDYLGKTV 111
Query 61 QVIPHITNEIKERIIR--------GGEGHDVVLVEVGGTVGDIESLPF 100
Q++PH+TN I++ I R G DV ++E+GGTVGDIES PF
Sbjct 112 QIVPHLTNAIQDWIERVAKIPVDDTGMEPDVCIIELGGTVGDIESAPF 159
> YJR103w
Length=564
Score = 127 bits (318), Expect = 6e-30, Method: Compositional matrix adjust.
Identities = 59/108 (54%), Positives = 80/108 (74%), Gaps = 8/108 (7%)
Query 1 PIQHGEVFVTDDGAETDLDLGHYERFIRTKMTRRNNFTTGRVYSEVLRKERRGDYLGATI 60
P++HGE FV DDG ETDLDLG+YER++ ++R +N TTG++YS V+ +ERRGDYLG T+
Sbjct 52 PLEHGECFVLDDGGETDLDLGNYERYLGITLSRDHNITTGKIYSHVISRERRGDYLGKTV 111
Query 61 QVIPHITNEIKERIIR--------GGEGHDVVLVEVGGTVGDIESLPF 100
Q++PH+TN I++ I R G DV ++E+GGTVGDIES PF
Sbjct 112 QIVPHLTNAIQDWIQRVSKIPVDDTGLEPDVCIIELGGTVGDIESAPF 159
> At2g34890
Length=597
Score = 126 bits (316), Expect = 1e-29, Method: Composition-based stats.
Identities = 59/108 (54%), Positives = 79/108 (73%), Gaps = 8/108 (7%)
Query 1 PIQHGEVFVTDDGAETDLDLGHYERFIRTKMTRRNNFTTGRVYSEVLRKERRGDYLGATI 60
P +HGEVFV DDG+E DLDLG+YERF+ T +TR NN T G++ V+ KER+GDYLG T+
Sbjct 52 PYEHGEVFVLDDGSEVDLDLGNYERFLDTTLTRDNNITYGKIQRYVMEKERKGDYLGETV 111
Query 61 QVIPHITNEIKERIIR-------GGEG-HDVVLVEVGGTVGDIESLPF 100
Q++PH+T+ I+E + R G EG DV ++E+GGT+GD ES PF
Sbjct 112 QIVPHVTDTIREWVERVAMIPVDGKEGPPDVCIIELGGTIGDNESRPF 159
> Hs21361829
Length=586
Score = 124 bits (310), Expect = 6e-29, Method: Composition-based stats.
Identities = 54/108 (50%), Positives = 76/108 (70%), Gaps = 8/108 (7%)
Query 1 PIQHGEVFVTDDGAETDLDLGHYERFIRTKMTRRNNFTTGRVYSEVLRKERRGDYLGATI 60
P +HGEVFV +DG E DLDLG+YERF+ + + NN TTG++Y V+ KERRGDYLG T+
Sbjct 52 PYEHGEVFVLNDGGEVDLDLGNYERFLDINLYKDNNITTGKIYQHVINKERRGDYLGKTV 111
Query 61 QVIPHITNEIKERIIRGG--------EGHDVVLVEVGGTVGDIESLPF 100
QV+PHIT+ ++E ++ E + ++E+GGT+GDIE +PF
Sbjct 112 QVVPHITDAVQEWVMNQAKVPVDGNKEEPQICVIELGGTIGDIEGMPF 159
> 7294316
Length=627
Score = 123 bits (309), Expect = 7e-29, Method: Compositional matrix adjust.
Identities = 57/107 (53%), Positives = 77/107 (71%), Gaps = 7/107 (6%)
Query 1 PIQHGEVFVTDDGAETDLDLGHYERFIRTKMTRRNNFTTGRVYSEVLRKERRGDYLGATI 60
P +HGEV+V DDGAE DLDLG+YERF+ + R NN TTG++Y V+ KER G+YLG T+
Sbjct 52 PYEHGEVYVLDDGAEVDLDLGNYERFLDVTLHRDNNITTGKIYKLVIEKERTGEYLGKTV 111
Query 61 QVIPHITNEIKERI-------IRGGEGHDVVLVEVGGTVGDIESLPF 100
QV+PHIT+ I+E + ++G V +VE+GGT+GDIE +PF
Sbjct 112 QVVPHITDAIQEWVERVAQTPVQGSSKPQVCIVELGGTIGDIEGMPF 158
> HsM9789919
Length=586
Score = 121 bits (304), Expect = 3e-28, Method: Compositional matrix adjust.
Identities = 54/108 (50%), Positives = 76/108 (70%), Gaps = 8/108 (7%)
Query 1 PIQHGEVFVTDDGAETDLDLGHYERFIRTKMTRRNNFTTGRVYSEVLRKERRGDYLGATI 60
P +HGEVFV +DG E DLDLG+YERF+ + + NN TTG++Y V+ KERRGDYLG T+
Sbjct 52 PYEHGEVFVLNDGGEVDLDLGNYERFLDINLYKDNNITTGKIYQHVINKERRGDYLGKTV 111
Query 61 QVIPHITNEIKERIIRGG--------EGHDVVLVEVGGTVGDIESLPF 100
QV+PHIT+ ++E ++ E + ++E+GGT+GDIE +PF
Sbjct 112 QVVPHITDAVQEWVMNQAKVPVDGNKEEPQICVIELGGTIGDIEGMPF 159
> CE19014
Length=599
Score = 117 bits (294), Expect = 4e-27, Method: Compositional matrix adjust.
Identities = 56/108 (51%), Positives = 77/108 (71%), Gaps = 8/108 (7%)
Query 1 PIQHGEVFVTDDGAETDLDLGHYERFIRTKMTRRNNFTTGRVYSEVLRKERRGDYLGATI 60
P +HGEVFV DDG E DLDLG+YERF+ ++TR NN TTG+++ V+ +ERRGDY G T+
Sbjct 58 PYEHGEVFVLDDGGEVDLDLGNYERFLDIRLTRDNNITTGKMFKHVMERERRGDYCGKTV 117
Query 61 QVIPHITNEIK---ERIIR-----GGEGHDVVLVEVGGTVGDIESLPF 100
Q+IPH+T+ I ER+ R E DV ++E+GGT+GDIE + +
Sbjct 118 QMIPHLTDAIVDWIERVARIPVDGTSEQPDVCIIELGGTIGDIEGMTY 165
> ECU11g0880
Length=535
Score = 90.9 bits (224), Expect = 6e-19, Method: Compositional matrix adjust.
Identities = 48/109 (44%), Positives = 69/109 (63%), Gaps = 12/109 (11%)
Query 1 PIQHGEVFVTDDGAETDLDLGHYERFIRTKMTRRNNFTTGRVYSEVLRKERRGDYLGATI 60
P +HGEV+V DDG E D+D G+YERF K++ N+ GR+ ++++ ER G +LG T+
Sbjct 52 PYEHGEVYVLDDGHECDMDFGNYERFNGIKLSGANSIPGGRLLHDIVKCEREGSFLGKTL 111
Query 61 QVIPHITNEIKERI----------IRGGEGH--DVVLVEVGGTVGDIES 97
Q+ PHI +E+ RI GG+ DVV+VE+GGTVG+ ES
Sbjct 112 QINPHIIDEVIRRIRAVADTPVESFGGGQAAVPDVVVVELGGTVGEYES 160
> ECU11g0480
Length=535
Score = 90.9 bits (224), Expect = 6e-19, Method: Compositional matrix adjust.
Identities = 48/109 (44%), Positives = 69/109 (63%), Gaps = 12/109 (11%)
Query 1 PIQHGEVFVTDDGAETDLDLGHYERFIRTKMTRRNNFTTGRVYSEVLRKERRGDYLGATI 60
P +HGEV+V DDG E D+D G+YERF K++ N+ GR+ ++++ ER G +LG T+
Sbjct 52 PYEHGEVYVLDDGHECDMDFGNYERFNGIKLSGANSIPGGRLLHDIVKCEREGSFLGKTL 111
Query 61 QVIPHITNEIKERI----------IRGGEGH--DVVLVEVGGTVGDIES 97
Q+ PHI +E+ RI GG+ DVV+VE+GGTVG+ ES
Sbjct 112 QINPHIIDEVIRRIRAVADTPVESFGGGQAAVPDVVVVELGGTVGEYES 160
> 7296017
Length=971
Score = 28.5 bits (62), Expect = 3.0, Method: Composition-based stats.
Identities = 11/24 (45%), Positives = 16/24 (66%), Gaps = 0/24 (0%)
Query 4 HGEVFVTDDGAETDLDLGHYERFI 27
HG VF+ DDGA+ ++L +R I
Sbjct 542 HGPVFIIDDGAQPKIELASKDRNI 565
> CE05460
Length=338
Score = 28.1 bits (61), Expect = 4.2, Method: Composition-based stats.
Identities = 15/45 (33%), Positives = 22/45 (48%), Gaps = 2/45 (4%)
Query 26 FIRTKMTRRNNFTTGRVYSEVLRKERRGDYLGATIQVIPHITNEI 70
F + +MT+ N TT E+ RK +L ++P ITN I
Sbjct 223 FYKLRMTQNN--TTSERTKELTRKAEYSLFLAVVSSIVPFITNGI 265
> CE26929
Length=440
Score = 27.7 bits (60), Expect = 5.5, Method: Composition-based stats.
Identities = 18/54 (33%), Positives = 28/54 (51%), Gaps = 3/54 (5%)
Query 17 DLDLGHYERFIR--TKMTRRNNFTTGRVYSEVLRKERRGDYLGATIQVIPHITN 68
D G+YE ++ + T + + +G YS L RRGDY A +++I I N
Sbjct 151 DYKEGNYEEALKKFNEATEFSGYQSGLAYSIALCHYRRGDYDSA-LKLISEIIN 203
> 7293781
Length=1339
Score = 27.3 bits (59), Expect = 7.0, Method: Composition-based stats.
Identities = 17/51 (33%), Positives = 28/51 (54%), Gaps = 2/51 (3%)
Query 37 FTTGRVYSEVLRKERRGDYLGATIQVIPHITNEIKER-IIRGGEGHDVVLV 86
F+ + Y L ++ + L Q + H+ + E+ ++RGGE HDVVLV
Sbjct 728 FSVSKAYGRALEQQIKTVELRLQRQHLEHVLAFVPEQFLLRGGE-HDVVLV 777
> Hs21536464
Length=585
Score = 27.3 bits (59), Expect = 7.3, Method: Composition-based stats.
Identities = 16/54 (29%), Positives = 28/54 (51%), Gaps = 0/54 (0%)
Query 45 EVLRKERRGDYLGATIQVIPHITNEIKERIIRGGEGHDVVLVEVGGTVGDIESL 98
+++R + + LGATI+ H + RI+RGG LV VG + ++ +
Sbjct 136 KIVRLVKNKEPLGATIRRDEHSGAVVVARIMRGGAADRSGLVHVGDELREVNGI 189
> HsM4505239
Length=585
Score = 27.3 bits (59), Expect = 7.4, Method: Composition-based stats.
Identities = 16/54 (29%), Positives = 28/54 (51%), Gaps = 0/54 (0%)
Query 45 EVLRKERRGDYLGATIQVIPHITNEIKERIIRGGEGHDVVLVEVGGTVGDIESL 98
+++R + + LGATI+ H + RI+RGG LV VG + ++ +
Sbjct 136 KIVRLVKNKEPLGATIRRDEHSGAVVVARIMRGGAADRSGLVHVGDELREVNGI 189
Lambda K H
0.320 0.143 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1187882580
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40