bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_1300_orf1
Length=110
Score E
Sequences producing significant alignments: (Bits) Value
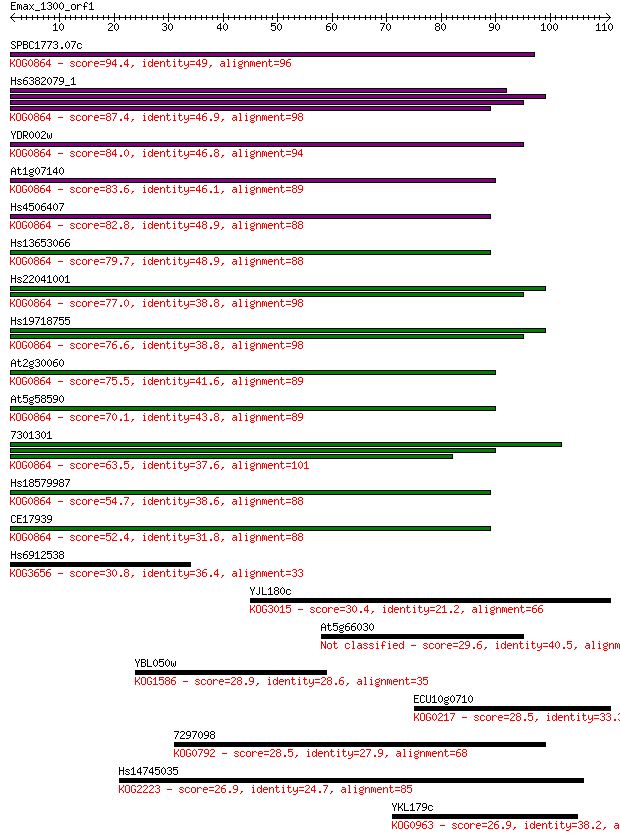
SPBC1773.07c 94.4 5e-20
Hs6382079_1 87.4 6e-18
YDR002w 84.0 7e-17
At1g07140 83.6 9e-17
Hs4506407 82.8 1e-16
Hs13653066 79.7 1e-15
Hs22041001 77.0 9e-15
Hs19718755 76.6 1e-14
At2g30060 75.5 2e-14
At5g58590 70.1 1e-12
7301301 63.5 8e-11
Hs18579987 54.7 5e-08
CE17939 52.4 2e-07
Hs6912538 30.8 0.68
YJL180c 30.4 0.80
At5g66030 29.6 1.4
YBL050w 28.9 2.2
ECU10g0710 28.5 3.1
7297098 28.5 3.6
Hs14745035 26.9 8.8
YKL179c 26.9 9.1
> SPBC1773.07c
Length=215
Score = 94.4 bits (233), Expect = 5e-20, Method: Compositional matrix adjust.
Identities = 47/97 (48%), Positives = 65/97 (67%), Gaps = 4/97 (4%)
Query 1 GEAKLLQHKETKRIRFLLRQEKTLKIVANHYVVATDVYCKLTPNVSSEKIWVWTV-MDFA 59
G+A+LL+HKET + R ++R++KTLK+ ANH ++ KLTPNV S++ WVWTV D +
Sbjct 119 GDARLLKHKETGKTRLVMRRDKTLKVCANHLLMP---EMKLTPNVGSDRSWVWTVAADVS 175
Query 60 EGELKNEQFALKFGQVEQAKEFKEKFEEAATINAKLF 96
EGE E FA++F E A FKE FE+ NAK+
Sbjct 176 EGEPTAETFAIRFANSENANLFKENFEKYQEENAKIL 212
> Hs6382079_1
Length=3065
Score = 87.4 bits (215), Expect = 6e-18, Method: Compositional matrix adjust.
Identities = 43/91 (47%), Positives = 63/91 (69%), Gaps = 3/91 (3%)
Query 1 GEAKLLQHKETKRIRFLLRQEKTLKIVANHYVVATDVYCKLTPNVSSEKIWVWTVMDFAE 60
G K+L+HK + +IR L+R+E+ LKI ANHY+ + D+ KLTPN S++ +VW +D+A+
Sbjct 1217 GNVKILRHKTSGKIRLLMRREQVLKICANHYI-SPDM--KLTPNAGSDRSFVWHALDYAD 1273
Query 61 GELKNEQFALKFGQVEQAKEFKEKFEEAATI 91
K EQ A++F E+A FK KFEEA +I
Sbjct 1274 ELPKPEQLAIRFKTPEEAALFKCKFEEAQSI 1304
Score = 77.0 bits (188), Expect = 7e-15, Method: Compositional matrix adjust.
Identities = 39/98 (39%), Positives = 60/98 (61%), Gaps = 6/98 (6%)
Query 1 GEAKLLQHKETKRIRFLLRQEKTLKIVANHYVVATDVYCKLTPNVSSEKIWVWTVMDFAE 60
G K+L+++ ++R L+R+E+ LK+ ANH++ T L P S++ W+W DF++
Sbjct 2058 GNLKILKNEVNGKLRMLMRREQVLKVCANHWITTT---MNLKPLSGSDRAWMWLASDFSD 2114
Query 61 GELKNEQFALKFGQVEQAKEFKEKFEEAATINAKLFDI 98
G+ K EQ A KF E A+EFK+KFEE + L DI
Sbjct 2115 GDAKLEQLAAKFKTPELAEEFKQKFEECQRL---LLDI 2149
Score = 72.4 bits (176), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 35/100 (35%), Positives = 62/100 (62%), Gaps = 15/100 (15%)
Query 1 GEAKLLQHKETKRIRFLLRQEKTLKIVANHYVVATDVYCKLTPNVS------SEKIWVWT 54
G+ K+LQ+ + K++R ++R+++ LK+ ANH ++TP+++ +E++W+WT
Sbjct 2355 GDIKILQNYDNKQVRIVMRRDQVLKLCANH---------RITPDMTLQNMKGTERVWLWT 2405
Query 55 VMDFAEGELKNEQFALKFGQVEQAKEFKEKFEEAATINAK 94
DFA+GE K E A++F + A FK+ F+EA T K
Sbjct 2406 ACDFADGERKVEHLAVRFKLQDVADSFKKIFDEAKTAQEK 2445
Score = 62.4 bits (150), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 36/89 (40%), Positives = 52/89 (58%), Gaps = 5/89 (5%)
Query 1 GEAKLLQHKETKRIRFLLRQEKTLKIVANHYVVATDVYCKLTP-NVSSEKIWVWTVMDFA 59
G+ K+L H R L+R+++ K+ ANH + T +L P NVS+ + VWT D+A
Sbjct 2956 GDIKILWHTMKNYYRILMRRDQVFKVCANHVITKT---MELKPLNVSNNAL-VWTASDYA 3011
Query 60 EGELKNEQFALKFGQVEQAKEFKEKFEEA 88
+GE K EQ A++F E A FK+ FEE
Sbjct 3012 DGEAKVEQLAVRFKTKEVADCFKKTFEEC 3040
> YDR002w
Length=201
Score = 84.0 bits (206), Expect = 7e-17, Method: Compositional matrix adjust.
Identities = 44/95 (46%), Positives = 65/95 (68%), Gaps = 4/95 (4%)
Query 1 GEAKLLQHKETKRIRFLLRQEKTLKIVANHYVVATDVYCKLTPNVSSEKIWVWTVM-DFA 59
G+ K L++K+T ++R L+R++KTLKI ANH ++A + L PNV S++ WV+ D A
Sbjct 109 GDCKFLKNKKTNKVRILMRRDKTLKICANH-IIAPEYT--LKPNVGSDRSWVYACTADIA 165
Query 60 EGELKNEQFALKFGQVEQAKEFKEKFEEAATINAK 94
EGE + FA++FG E A +FKE+FE+A IN K
Sbjct 166 EGEAEAFTFAIRFGSKENADKFKEEFEKAQEINKK 200
> At1g07140
Length=228
Score = 83.6 bits (205), Expect = 9e-17, Method: Compositional matrix adjust.
Identities = 41/89 (46%), Positives = 57/89 (64%), Gaps = 3/89 (3%)
Query 1 GEAKLLQHKETKRIRFLLRQEKTLKIVANHYVVATDVYCKLTPNVSSEKIWVWTVMDFAE 60
G K L+HK T +IR ++RQ KTLKI ANH+V + + +V +EK VW DFA+
Sbjct 72 GTVKFLKHKNTGKIRLVMRQSKTLKICANHFVKSG---MSVQEHVGNEKSCVWHARDFAD 128
Query 61 GELKNEQFALKFGQVEQAKEFKEKFEEAA 89
GELK+E F ++F +E K F +KF+E A
Sbjct 129 GELKDELFCIRFASIENCKTFMQKFKEVA 157
> Hs4506407
Length=201
Score = 82.8 bits (203), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 43/89 (48%), Positives = 57/89 (64%), Gaps = 4/89 (4%)
Query 1 GEAKLLQHKETKRIRFLLRQEKTLKIVANHYVVATDVYCKLTPNVSSEKIWVW-TVMDFA 59
G+ KLL+HKE IR L+R++KTLKI ANHY+ +L PN S++ WVW T DFA
Sbjct 73 GDVKLLKHKEKGAIRLLMRRDKTLKICANHYITP---MMELKPNAGSDRAWVWNTHADFA 129
Query 60 EGELKNEQFALKFGQVEQAKEFKEKFEEA 88
+ K E A++F E A++FK KFEE
Sbjct 130 DECPKPELLAIRFLNAENAQKFKTKFEEC 158
> Hs13653066
Length=201
Score = 79.7 bits (195), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 43/89 (48%), Positives = 57/89 (64%), Gaps = 4/89 (4%)
Query 1 GEAKLLQHKETKRIRFLLRQEKTLKIVANHYVVATDVYCKLTPNVSSEKIWVW-TVMDFA 59
G+ KLL+HKE IR L++++KTLKI ANHY T +L PN S++ WVW T DFA
Sbjct 73 GDVKLLKHKEKGAIRLLMQRDKTLKICANHY---TTPMMELKPNAGSDRAWVWNTHADFA 129
Query 60 EGELKNEQFALKFGQVEQAKEFKEKFEEA 88
+ K E A++F E A++FK KFEE
Sbjct 130 DECPKPELLAIRFLNAENAQKFKTKFEEC 158
> Hs22041001
Length=1021
Score = 77.0 bits (188), Expect = 9e-15, Method: Compositional matrix adjust.
Identities = 38/98 (38%), Positives = 61/98 (62%), Gaps = 6/98 (6%)
Query 1 GEAKLLQHKETKRIRFLLRQEKTLKIVANHYVVATDVYCKLTPNVSSEKIWVWTVMDFAE 60
G K+L+++ ++R L+R+E+ LK+ ANH++ T L P S++ W+W+ DF++
Sbjct 338 GNLKILKNEVNGKLRMLMRREQVLKVCANHWITTT---MNLKPLSGSDRAWMWSASDFSD 394
Query 61 GELKNEQFALKFGQVEQAKEFKEKFEEAATINAKLFDI 98
G+ K E+ A KF E A+EFK+KFEE + L DI
Sbjct 395 GDAKLERLAAKFKTPELAEEFKQKFEECQRL---LLDI 429
Score = 74.3 bits (181), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 37/100 (37%), Positives = 62/100 (62%), Gaps = 15/100 (15%)
Query 1 GEAKLLQHKETKRIRFLLRQEKTLKIVANHYVVATDVYCKLTPNVS------SEKIWVWT 54
G+ K+LQ+ + K++R ++R+++ LK+ ANH ++TP++S +E++WVWT
Sbjct 635 GDIKILQNYDNKQVRIVMRRDQVLKLCANH---------RITPDMSLQNMKGTERVWVWT 685
Query 55 VMDFAEGELKNEQFALKFGQVEQAKEFKEKFEEAATINAK 94
DFA+GE K E A++F + A FK+ F+EA T K
Sbjct 686 ACDFADGERKVEHLAVRFKLQDVADSFKKIFDEAKTAQEK 725
> Hs19718755
Length=1765
Score = 76.6 bits (187), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 38/98 (38%), Positives = 61/98 (62%), Gaps = 6/98 (6%)
Query 1 GEAKLLQHKETKRIRFLLRQEKTLKIVANHYVVATDVYCKLTPNVSSEKIWVWTVMDFAE 60
G K+L+++ ++R L+R+E+ LK+ ANH++ T L P S++ W+W+ DF++
Sbjct 1082 GNLKILKNEVNGKLRMLMRREQVLKVCANHWITTT---MNLKPLSGSDRAWMWSASDFSD 1138
Query 61 GELKNEQFALKFGQVEQAKEFKEKFEEAATINAKLFDI 98
G+ K E+ A KF E A+EFK+KFEE + L DI
Sbjct 1139 GDAKLERLAAKFKTPELAEEFKQKFEECQRL---LLDI 1173
Score = 74.3 bits (181), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 37/100 (37%), Positives = 62/100 (62%), Gaps = 15/100 (15%)
Query 1 GEAKLLQHKETKRIRFLLRQEKTLKIVANHYVVATDVYCKLTPNVS------SEKIWVWT 54
G+ K+LQ+ + K++R ++R+++ LK+ ANH ++TP++S +E++WVWT
Sbjct 1379 GDIKILQNYDNKQVRIVMRRDQVLKLCANH---------RITPDMSLQNMKGTERVWVWT 1429
Query 55 VMDFAEGELKNEQFALKFGQVEQAKEFKEKFEEAATINAK 94
DFA+GE K E A++F + A FK+ F+EA T K
Sbjct 1430 ACDFADGERKVEHLAVRFKLQDVADSFKKIFDEAKTAQEK 1469
> At2g30060
Length=217
Score = 75.5 bits (184), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 37/89 (41%), Positives = 55/89 (61%), Gaps = 3/89 (3%)
Query 1 GEAKLLQHKETKRIRFLLRQEKTLKIVANHYVVATDVYCKLTPNVSSEKIWVWTVMDFAE 60
G K L+H+ + +IR ++RQ KTLKI ANH V + + + ++K VW DF++
Sbjct 74 GTVKFLKHRVSGKIRLVMRQSKTLKICANHLVGSG---MSVQEHAGNDKSCVWHARDFSD 130
Query 61 GELKNEQFALKFGQVEQAKEFKEKFEEAA 89
GELK+E F ++F VE K F +KF+E A
Sbjct 131 GELKDELFCIRFASVENCKAFMQKFKEVA 159
> At5g58590
Length=260
Score = 70.1 bits (170), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 39/107 (36%), Positives = 57/107 (53%), Gaps = 21/107 (19%)
Query 1 GEAKLLQHKETKRIRFLLRQEKTLKIVANHYVVATDVYCKLTPNVSSEKIWVWTVMDFAE 60
G KLL+HKET ++R ++RQ KTLKI ANH + + + + +EK +W DF++
Sbjct 84 GTVKLLKHKETGKVRLVMRQSKTLKICANHLISSG---MSVQEHSGNEKSCLWHATDFSD 140
Query 61 GELKNEQFALKFGQVEQ------------------AKEFKEKFEEAA 89
GELK+E F ++F +E+ K F EKF E A
Sbjct 141 GELKDELFCIRFASIERKMVWKILEWLTDSVASTDCKTFMEKFTEIA 187
> 7301301
Length=2659
Score = 63.5 bits (153), Expect = 8e-11, Method: Compositional matrix adjust.
Identities = 38/101 (37%), Positives = 59/101 (58%), Gaps = 7/101 (6%)
Query 1 GEAKLLQHKETKRIRFLLRQEKTLKIVANHYVVATDVYCKLTPNVSSEKIWVWTVMDFAE 60
G+ K+L+H++TK++R ++R+E+ KI NH + VY + T E W++ V DF+E
Sbjct 2027 GDVKILRHRQTKKLRVVMRREQVFKICLNHVLNENVVYREKT-----ETSWMFAVHDFSE 2081
Query 61 GELKNEQFALKFGQVEQAKEFKEKFEEAATINAKLFDIEES 101
GE E+F L+F E A+ F E + A AK IE+S
Sbjct 2082 GESVLERFTLRFKNKEVAQGFMEAIKNALNETAK--PIEDS 2120
Score = 50.8 bits (120), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 29/89 (32%), Positives = 50/89 (56%), Gaps = 1/89 (1%)
Query 1 GEAKLLQHKETKRIRFLLRQEKTLKIVANHYVVATDVYCKLTPNVSSEKIWVWTVMDFAE 60
G K+L K T R L+R+++T K+ ANH + A D+ + +K +W DFA+
Sbjct 1317 GVIKILCDKATGVSRVLMRRDQTHKVCANHTITA-DITINVANQDKDKKSLLWAANDFAD 1375
Query 61 GELKNEQFALKFGQVEQAKEFKEKFEEAA 89
++ E+F ++F E A+EF+ F +A+
Sbjct 1376 EQVTLERFLVRFKTGELAEEFRVAFTKAS 1404
Score = 37.4 bits (85), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 26/88 (29%), Positives = 47/88 (53%), Gaps = 10/88 (11%)
Query 1 GEAKLLQHKETKRIRFLLRQEKTLKIVANHYVVATDVYCKLTPNVSSEKIWVWTVMDF-- 58
GE K+L+H E + R ++RQE+ K+V N + A+ ++ + K ++W ++
Sbjct 2543 GEIKVLEHPELQTFRLIMRQEQIHKLVLNMNISAS---LQMDYMNAQMKSFLWAGYNYAV 2599
Query 59 -AEGELKN----EQFALKFGQVEQAKEF 81
AEG++ E+ A +F + E A EF
Sbjct 2600 DAEGKVDTEGVLERLACRFAKEEIASEF 2627
> Hs18579987
Length=147
Score = 54.7 bits (130), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 34/88 (38%), Positives = 47/88 (53%), Gaps = 3/88 (3%)
Query 1 GEAKLLQHKETKRIRFLLRQEKTLKIVANHYVVATDVYCKLTPNVSSEKIWVWTVMDFAE 60
G+ KLL+HKE IR L++++KTL I ANHY++ +L P T M +
Sbjct 20 GDVKLLKHKEKGAIRLLMQRDKTLNICANHYIMR---MMELKPKQVVTVPGSGTPMLTSP 76
Query 61 GELKNEQFALKFGQVEQAKEFKEKFEEA 88
K E A+ F E A++FK KFEE
Sbjct 77 TIPKPELLAICFLNAENAQKFKTKFEEC 104
> CE17939
Length=860
Score = 52.4 bits (124), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 28/88 (31%), Positives = 52/88 (59%), Gaps = 3/88 (3%)
Query 1 GEAKLLQHKETKRIRFLLRQEKTLKIVANHYVVATDVYCKLTPNVSSEKIWVWTVMDFAE 60
GE K+L +K+ K R ++R+++ LK+ AN ++ + ++ S+EK + W DF+E
Sbjct 300 GELKVLYNKDKKSWRVVMRRDQVLKVCANFPILGSMTIQQMK---SNEKAYTWFCEDFSE 356
Query 61 GELKNEQFALKFGQVEQAKEFKEKFEEA 88
+ + + + +F V+ A EFK FE+A
Sbjct 357 DQPAHVKLSARFANVDIAGEFKTLFEKA 384
> Hs6912538
Length=410
Score = 30.8 bits (68), Expect = 0.68, Method: Composition-based stats.
Identities = 12/33 (36%), Positives = 21/33 (63%), Gaps = 0/33 (0%)
Query 1 GEAKLLQHKETKRIRFLLRQEKTLKIVANHYVV 33
G+ L++HK+ +RIR L R + L+ + YV+
Sbjct 276 GQVSLVRHKDVRRIRSLQRSVQVLRAIVVMYVI 308
> YJL180c
Length=325
Score = 30.4 bits (67), Expect = 0.80, Method: Compositional matrix adjust.
Identities = 14/66 (21%), Positives = 34/66 (51%), Gaps = 0/66 (0%)
Query 45 VSSEKIWVWTVMDFAEGELKNEQFALKFGQVEQAKEFKEKFEEAATINAKLFDIEESGSK 104
+ ++ + V++ M+ EG L+N Q L ++ +EF F + I ++ D + G +
Sbjct 159 LDTDTLLVFSPMNEFEGRLRNAQNELYIPIIKGMEEFLRNFSSESNIRLQILDADIHGLR 218
Query 105 EGEKAE 110
++++
Sbjct 219 GNQQSD 224
> At5g66030
Length=767
Score = 29.6 bits (65), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 15/39 (38%), Positives = 28/39 (71%), Gaps = 2/39 (5%)
Query 58 FAEGELKNEQFALKFGQVEQA--KEFKEKFEEAATINAK 94
++E + K+++++ KF QVEQ +E KE+ E+ A ++AK
Sbjct 119 YSEADAKSQEYSSKFSQVEQKLDQEIKERDEKYADLDAK 157
> YBL050w
Length=292
Score = 28.9 bits (63), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 10/35 (28%), Positives = 20/35 (57%), Gaps = 0/35 (0%)
Query 24 LKIVANHYVVATDVYCKLTPNVSSEKIWVWTVMDF 58
LK + Y+ A+D+Y KL + ++ W++ D+
Sbjct 167 LKALDGQYIEASDIYSKLIKSSMGNRLSQWSLKDY 201
> ECU10g0710
Length=922
Score = 28.5 bits (62), Expect = 3.1, Method: Composition-based stats.
Identities = 12/36 (33%), Positives = 23/36 (63%), Gaps = 0/36 (0%)
Query 75 VEQAKEFKEKFEEAATINAKLFDIEESGSKEGEKAE 110
VE+A+++ + EE + K++D+ E+G GE+ E
Sbjct 528 VEKAEDYSCRIEEVLVWHRKVYDVTEAGISPGEENE 563
> 7297098
Length=1252
Score = 28.5 bits (62), Expect = 3.6, Method: Composition-based stats.
Identities = 19/74 (25%), Positives = 34/74 (45%), Gaps = 6/74 (8%)
Query 31 YVVATDVYCKLTPNVSSEKIW---VWTVMDFAEGEL---KNEQFALKFGQVEQAKEFKEK 84
Y+VA LT + + +W V+ V+ E +N L+FGQ + +EF +
Sbjct 1053 YIVAQSPQEPLTMRIFWQCVWEADVYLVVQLTEDMSYIPRNSHQRLEFGQFQVYQEFSQT 1112
Query 85 FEEAATINAKLFDI 98
+ TI +L+ +
Sbjct 1113 TDRCTTIKLRLYHV 1126
> Hs14745035
Length=775
Score = 26.9 bits (58), Expect = 8.8, Method: Composition-based stats.
Identities = 21/85 (24%), Positives = 39/85 (45%), Gaps = 1/85 (1%)
Query 21 EKTLKIVANHYVVATDVYCKLTPNVSSEKIWVWTVMDFAEGELKNEQFALKFGQVEQAKE 80
+KT KI+ Y T CK P S K + + + L++ L VE+A
Sbjct 357 QKTSKIIQQEYEARTGRTCKPPPQSSRRKNFEFEPLSTTALILEDRPSNLPAKSVEEALR 416
Query 81 FKEKFEEAATINAKLFDIEESGSKE 105
+++++E AK +I+E+ ++
Sbjct 417 HRQEYDEMVA-EAKKREIKEAHKRK 440
> YKL179c
Length=679
Score = 26.9 bits (58), Expect = 9.1, Method: Compositional matrix adjust.
Identities = 13/44 (29%), Positives = 25/44 (56%), Gaps = 10/44 (22%)
Query 71 KFGQVEQAKEFKE----------KFEEAATINAKLFDIEESGSK 104
K G+++ +KE KE K+ + T+ ++L D+E+S +K
Sbjct 113 KLGKIDDSKELKEKISYLEDKLAKYADYETLKSRLLDLEQSSAK 156
Lambda K H
0.313 0.129 0.360
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1195973986
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40